Association of ATRX with pericentric heterochromatin and the Y chromosome of neonatal mouse spermatogonia
- PMID: 18366812
- PMCID: PMC2275742
- DOI: 10.1186/1471-2199-9-29
Association of ATRX with pericentric heterochromatin and the Y chromosome of neonatal mouse spermatogonia
Abstract
Background: Establishment of chromosomal cytosine methylation and histone methylation patterns are critical epigenetic modifications required for heterochromatin formation in the mammalian genome. However, the nature of the primary signal(s) targeting DNA methylation at specific genomic regions is not clear. Notably, whether histone methylation and/or chromatin remodeling proteins play a role in the establishment of DNA methylation during gametogenesis is not known. The chromosomes of mouse neonatal spermatogonia display a unique pattern of 5-methyl cytosine staining whereby centromeric heterochromatin is hypo-methylated whereas chromatids are strongly methylated. Thus, in order to gain some insight into the relationship between global DNA and histone methylation in the germ line we have used neonatal spermatogonia as a model to determine whether these unique chromosomal DNA methylation patterns are also reflected by concomitant changes in histone methylation.
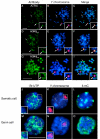
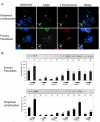
Results: Our results demonstrate that histone H3 tri-methylated at lysine 9 (H3K9me3), a hallmark of constitutive heterochromatin, as well as the chromatin remodeling protein ATRX remained associated with pericentric heterochromatin regions in spite of their extensive hypo-methylation. This suggests that in neonatal spermatogonia, chromosomal 5-methyl cytosine patterns are regulated independently of changes in histone methylation, potentially reflecting a crucial mechanism to maintain pericentric heterochromatin silencing. Furthermore, chromatin immunoprecipitation and fluorescence in situ hybridization, revealed that ATRX as well as H3K9me3 associate with Y chromosome-specific DNA sequences and decorate both arms of the Y chromosome, suggesting a possible role in heterochromatinization and the predominant transcriptional quiescence of this chromosome during spermatogenesis.
Conclusion: These results are consistent with a role for histone modifications and chromatin remodeling proteins such as ATRX in maintaining transcriptional repression at constitutive heterochromatin domains in the absence of 5-methyl cytosine and provide evidence suggesting that the establishment and/or maintenance of repressive histone and chromatin modifications at pericentric heterochromatin following genome-wide epigenetic reprogramming in the germ line may precede the establishment of chromosomal 5-methyl cytosine patterns as a genomic silencing strategy in neonatal spermatogonia.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

