Masking of the endoplasmic reticulum retention signals during assembly of the NMDA receptor
- PMID: 18367616
- PMCID: PMC6670593
- DOI: 10.1523/JNEUROSCI.5239-07.2008
Masking of the endoplasmic reticulum retention signals during assembly of the NMDA receptor
Abstract
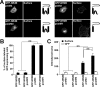
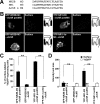
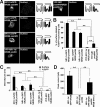
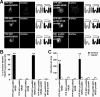
NMDA receptors are glutamate-gated ion channels that play important roles in synaptic transmission and excitotoxicity. The functional NMDA receptor is thought to be a heterotetramer composed mainly of two NR1 and two NR2 subunits. Although it is generally accepted that only correctly assembled NMDA receptors can pass the ER quality control, the mechanism underlying this process is not well understood. Using truncated and chimeric NMDA receptor subunits expressed in heterologous cells and cortical neurons, we found that the third membrane domains (M3) of both NR1 and NR2B contain signals that cause the unassembled subunits to be retained in the ER. M3 of both NR1 and NR2B and, M4 of NR1, are necessary for masking ER retention signals found in M3. Thus, our data reveal a critical role of the membrane domains in the assembly of functional NMDA receptors.
Figures





References
-
- Ayalon G, Stern-Bach Y. Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron. 2001;31:103–113. - PubMed
-
- Call ME, Wucherpfennig KW. The T cell receptor: critical role of the membrane environment in receptor assembly and function. Annu Rev Immunol. 2005;23:101–125. - PubMed
-
- Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. - PubMed
-
- Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
