Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda
- PMID: 18369368
- PMCID: PMC2278112
- DOI: 10.1038/embor.2008.33
Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda
Abstract
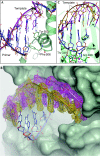
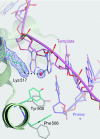
The simple deletion of nucleotides is common in many organisms. It can be advantageous when it activates genes beneficial to microbial survival in adverse environments, and deleterious when it mutates genes relevant to survival, cancer or degenerative diseases. The classical idea is that simple deletions arise by strand slippage. A prime opportunity for slippage occurs during DNA synthesis, but it remains unclear how slippage is controlled during a polymerization cycle. Here, we report crystal structures and molecular dynamics simulations of mutant derivatives of DNA polymerase lambda bound to a primer-template during strand slippage. Relative to the primer strand, the template strand is in multiple conformations, indicating intermediates on the pathway to deletion mutagenesis. Consistent with these intermediates, the mutant polymerases generate single-base deletions at high rates. The results indicate that dNTP-induced template strand repositioning during conformational rearrangements in the catalytic cycle is crucial to controlling the rate of strand slippage.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Bebenek K, Kunkel TA (2000) Streisinger revisited: DNA synthesis errors mediated by substrate misalignments. Cold Spring Harb Symp Quant Biol 65: 81–91 - PubMed
-
- Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA (2003) The frameshift infidelity of human DNA polymerase λ. Implications for function. J Biol Chem 278: 34685–34690 - PubMed
-
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) Charmm: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4: 187–217
-
- Brünger AT et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 - PubMed
-
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

