Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics
- PMID: 18369420
- PMCID: PMC2265556
- DOI: 10.1371/journal.pcbi.1000021
Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics
Erratum in
- PLoS Comput Biol. 2008 Mar;4(3). doi: 10.1371/annotation/1c55be5f-ecd7-49be-91c1-91881be60297
- PLoS Comput Biol. 2008 Mar;4(3). doi: 10.1371/annotation/e14ad837-e5ff-4bd5-a5f2-f32e784d75a2
Abstract
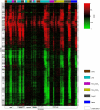
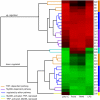
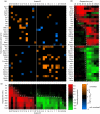
Macrophages are versatile immune cells that can detect a variety of pathogen-associated molecular patterns through their Toll-like receptors (TLRs). In response to microbial challenge, the TLR-stimulated macrophage undergoes an activation program controlled by a dynamically inducible transcriptional regulatory network. Mapping a complex mammalian transcriptional network poses significant challenges and requires the integration of multiple experimental data types. In this work, we inferred a transcriptional network underlying TLR-stimulated murine macrophage activation. Microarray-based expression profiling and transcription factor binding site motif scanning were used to infer a network of associations between transcription factor genes and clusters of co-expressed target genes. The time-lagged correlation was used to analyze temporal expression data in order to identify potential causal influences in the network. A novel statistical test was developed to assess the significance of the time-lagged correlation. Several associations in the resulting inferred network were validated using targeted ChIP-on-chip experiments. The network incorporates known regulators and gives insight into the transcriptional control of macrophage activation. Our analysis identified a novel regulator (TGIF1) that may have a role in macrophage activation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
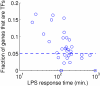

 (see Equation 4) as a function of the time lag τ, for Irf7 and Stat1. The peak value of ρτ
2 occurs at τ = 10, but the peak significance value (taking into account the lag-specific null distribution) occurs at τ = 20 min.
(see Equation 4) as a function of the time lag τ, for Irf7 and Stat1. The peak value of ρτ
2 occurs at τ = 10, but the peak significance value (taking into account the lag-specific null distribution) occurs at τ = 20 min.
References
-
- Underhill DM, Ozinsky A. Toll-like receptors: key mediators of microbe detection. Curr Opin Immunol. 2002;14:103–110. - PubMed
-
- Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. - PubMed
-
- Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. - PubMed
-
- Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. - PubMed
-
- Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, et al. Targeted disruption of the MyD88 gene results in loss of IL-1 and IL-18-mediated function. Immunity. 1998;9:143–150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

