GADD45A does not promote DNA demethylation
- PMID: 18369439
- PMCID: PMC2265528
- DOI: 10.1371/journal.pgen.1000013
GADD45A does not promote DNA demethylation
Abstract
Although DNA methylation patterns in somatic cells are thought to be relatively stable, they undergo dramatic changes during embryonic development, gametogenesis, and during malignant transformation. The enzymology of DNA methyltransferases is well understood, but the mechanism that removes methylated cytosines from DNA (active DNA demethylation) has remained enigmatic. Recently, a role of the growth arrest and DNA damage inducible protein GADD45A in DNA demethylation has been reported [1]. We have investigated the function of GADD45A in DNA demethylation in more detail using gene reactivation and DNA methylation assays. Contrary to the previous report, we were unable to substantiate a functional role of GADD45A in DNA demethylation. The mechanism of active DNA demethylation in mammalian cells remains unknown.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
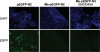


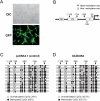


Similar articles
-
Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation.Nature. 2007 Feb 8;445(7128):671-5. doi: 10.1038/nature05515. Epub 2007 Jan 31. Nature. 2007. PMID: 17268471
-
Retinoic acid inhibits white adipogenesis by disrupting GADD45A-mediated Zfp423 DNA demethylation.J Mol Cell Biol. 2017 Aug 1;9(4):338-349. doi: 10.1093/jmcb/mjx026. J Mol Cell Biol. 2017. PMID: 28992291 Free PMC article.
-
GADD45a physically and functionally interacts with TET1.Differentiation. 2015 Jul-Oct;90(1-3):59-68. doi: 10.1016/j.diff.2015.10.003. Epub 2015 Nov 3. Differentiation. 2015. PMID: 26546041 Free PMC article.
-
Gadd45 proteins: key players of repair-mediated DNA demethylation.Adv Exp Med Biol. 2013;793:35-50. doi: 10.1007/978-1-4614-8289-5_3. Adv Exp Med Biol. 2013. PMID: 24104472 Review.
-
Terminal differentiation induction as DNA damage response in hematopoietic stem cells by GADD45A.Exp Hematol. 2016 Jul;44(7):561-6. doi: 10.1016/j.exphem.2016.04.006. Epub 2016 Jun 1. Exp Hematol. 2016. PMID: 27262218 Review.
Cited by
-
Imprinting and epigenetic changes in the early embryo.Mamm Genome. 2009 Sep-Oct;20(9-10):532-43. doi: 10.1007/s00335-009-9225-2. Epub 2009 Sep 16. Mamm Genome. 2009. PMID: 19760320 Review.
-
Analysis of TET expression/activity and 5mC oxidation during normal and malignant germ cell development.PLoS One. 2013 Dec 26;8(12):e82881. doi: 10.1371/journal.pone.0082881. eCollection 2013. PLoS One. 2013. PMID: 24386123 Free PMC article.
-
Gadd45a inhibits cell migration and invasion by altering the global RNA expression.Cancer Biol Ther. 2012 Sep;13(11):1112-22. doi: 10.4161/cbt.21186. Epub 2012 Jul 24. Cancer Biol Ther. 2012. PMID: 22825327 Free PMC article.
-
Reprogramming towards pluripotency requires AID-dependent DNA demethylation.Nature. 2010 Feb 25;463(7284):1042-7. doi: 10.1038/nature08752. Nature. 2010. PMID: 20027182 Free PMC article.
-
GADD45α inhibition of DNMT1 dependent DNA methylation during homology directed DNA repair.Nucleic Acids Res. 2012 Mar;40(6):2481-93. doi: 10.1093/nar/gkr1115. Epub 2011 Dec 1. Nucleic Acids Res. 2012. PMID: 22135303 Free PMC article.
References
-
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. - PubMed
-
- Pfeifer GP, Steigerwald SD, Hansen RS, Gartler SM, Riggs AD. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci U S A. 1990;87:8252–8256. - PMC - PubMed
-
- Paroush Z, Keshet I, Yisraeli J, Cedar H. Dynamics of demethylation and activation of the alpha-actin gene in myoblasts. Cell. 1990;63:1229–1237. - PubMed
-
- Saluz HP, Jiricny J, Jost JP. Genomic sequencing reveals a positive correlation between the kinetics of strand-specific DNA demethylation of the overlapping estradiol/glucocorticoid-receptor binding sites and the rate of avian vitellogenin mRNA synthesis. Proc Natl Acad Sci U S A. 1986;83:7167–7171. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

