From classical genetics to quantitative genetics to systems biology: modeling epistasis
- PMID: 18369448
- PMCID: PMC2265472
- DOI: 10.1371/journal.pgen.1000029
From classical genetics to quantitative genetics to systems biology: modeling epistasis
Erratum in
- PLoS Genet. 2008 May;4(5). doi: 10.1371/annotation/ff93eba8-9567-4f41-b90d-9cdfdf65f747 doi: 10.1371/annotation/ff93eba8-9567-4f41-b90d-9cdfdf65f747
Abstract
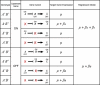
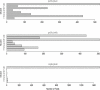
Gene expression data has been used in lieu of phenotype in both classical and quantitative genetic settings. These two disciplines have separate approaches to measuring and interpreting epistasis, which is the interaction between alleles at different loci. We propose a framework for estimating and interpreting epistasis from a classical experiment that combines the strengths of each approach. A regression analysis step accommodates the quantitative nature of expression measurements by estimating the effect of gene deletions plus any interaction. Effects are selected by significance such that a reduced model describes each expression trait. We show how the resulting models correspond to specific hierarchical relationships between two regulator genes and a target gene. These relationships are the basic units of genetic pathways and genomic system diagrams. Our approach can be extended to analyze data from a variety of experiments, multiple loci, and multiple environments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. - PubMed
-
- Lynch M, Walsh B. Genetics and the Analysis of Quantitative Traits. Sunderland, MA: Sinauer Associates; 1998.
-
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. - PubMed
-
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

