Recruitment to the nuclear periphery can alter expression of genes in human cells
- PMID: 18369458
- PMCID: PMC2265557
- DOI: 10.1371/journal.pgen.1000039
Recruitment to the nuclear periphery can alter expression of genes in human cells
Abstract
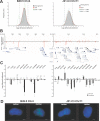
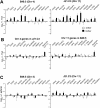
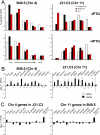
The spatial organisation of the genome in the nucleus has a role in the regulation of gene expression. In vertebrates, chromosomal regions with low gene-density are located close to the nuclear periphery. Correlations have also been made between the transcriptional state of some genes and their location near the nuclear periphery. However, a crucial issue is whether this level of nuclear organisation directly affects gene function, rather than merely reflecting it. To directly investigate whether proximity to the nuclear periphery can influence gene expression in mammalian cells, here we relocate specific human chromosomes to the nuclear periphery by tethering them to a protein of the inner nuclear membrane. We show that this can reversibly suppress the expression of some endogenous human genes located near the tethering sites, and even genes further away. However, the expression of many other genes is not detectably reduced and we show that location at the nuclear periphery is not incompatible with active transcription. The dampening of gene expression around the nuclear periphery is dependent on the activity of histone deacetylases. Our data show that the radial position within the nucleus can influence the expression of some, but not all, genes. This is compatible with the suggestion that re-localisation of genes relative to the peripheral zone of the nucleus could be used by metazoans to modulate the expression of selected genes during development and differentiation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, et al. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. - PubMed
-
- Cremer M, von Hase J, Volm T, Brero A, Kreth G, et al. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–567. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

