Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage
- PMID: 18369479
- PMCID: PMC2265427
- DOI: 10.1371/journal.ppat.1000033
Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage
Abstract
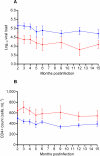
One of the most important genetic factors known to affect the rate of disease progression in HIV-infected individuals is the genotype at the Class I Human Leukocyte Antigen (HLA) locus, which determines the HIV peptides targeted by cytotoxic T-lymphocytes (CTLs). Individuals with HLA-B*57 or B*5801 alleles, for example, target functionally important parts of the Gag protein. Mutants that escape these CTL responses may have lower fitness than the wild-type and can be associated with slower disease progression. Transmission of the escape variant to individuals without these HLA alleles is associated with rapid reversion to wild-type. However, the question of whether infection with an escape mutant offers an advantage to newly infected hosts has not been addressed. Here we investigate the relationship between the genotypes of transmitted viruses and prognostic markers of disease progression and show that infection with HLA-B*57/B*5801 escape mutants is associated with lower viral load and higher CD4+ counts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Phillips RE, Rowland-Jones S, Nixon DF, Gotch FM, Edwards JP, et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354(6353):453–459. - PubMed
-
- Goulder P, Price D, Nowak M, Rowland-Jones S, Phillips R, et al. Co-evolution of human immunodeficiency virus and cytotoxic T-lymphocyte responses. Immunol Rev. 1997;159:17–29. - PubMed
-
- Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, et al. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3(2):205–211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

