The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication
- PMID: 18369482
- PMCID: PMC2275779
- DOI: 10.1371/journal.ppat.1000046
The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication
Abstract
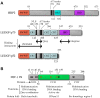
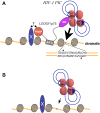
Retroviral replication proceeds through a stable proviral DNA intermediate, and numerous host cell factors have been implicated in its formation. In particular, recent results have highlighted an important role for the integrase-interactor lens epithelium-derived growth factor (LEDGF)/p75 in lentiviral integration. Cells engineered to over-express fragments of LEDGF/p75 containing its integrase-binding domain but lacking determinants essential for chromatin association are refractory to HIV-1 infection. Furthermore, both the levels of HIV-1 integration and the genomic distribution of the resultant proviruses are significantly perturbed in cells devoid of endogenous LEDGF/p75 protein. A strong bias towards integration along transcription units is a characteristic feature of lentiviruses. In the absence of LEDGF/p75, HIV-1 in large part loses that preference, displaying concomitant integration surges in the vicinities of CpG islands and gene promoter regions, elements naturally targeted by other types of retroviruses. Together, these findings highlight that LEDGF/p75 is an important albeit not strictly essential cofactor of lentiviral DNA integration, and solidify a role for chromatin-associated LEDGF/p75 as a receptor for lentiviral preintegration complexes. By now one of the best characterized virus-host interactions, the integrase-LEDGF/p75 interface opens a range of opportunities for lentiviral vector targeting for gene therapy applications as well as for the development of novel classes of antiretroviral drugs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

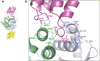
References
-
- Bowerman B, Brown PO, Bishop JM, Varmus HE. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. - PubMed
-
- Pauza C. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology. 1990;179:886–889. - PubMed
-
- Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: Mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. - PubMed
-
- Turlure F, Devroe E, Silver PA, Engelman A. Human cell proteins and human immunodeficiency virus DNA integration. Front Biosci. 2004;9:3187–3208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

