Comprehensive characterization of genomic aberrations in gangliogliomas by CGH, array-based CGH and interphase FISH
- PMID: 18371186
- PMCID: PMC8095656
- DOI: 10.1111/j.1750-3639.2008.00122.x
Comprehensive characterization of genomic aberrations in gangliogliomas by CGH, array-based CGH and interphase FISH
Abstract
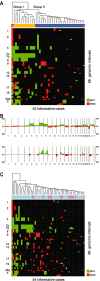
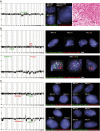
Gangliogliomas are generally benign neuroepithelial tumors composed of dysplastic neuronal and neoplastic glial elements. We screened 61 gangliogliomas [World Health Organization (WHO) grade I] for genomic alterations by chromosomal and array-based comparative genomic hybridization (CGH). Aberrations were detected in 66% of gangliogliomas (mean +/- SEM = 2.5 +/- 0.5 alterations/tumor). Frequent gains were on chromosomes 7 (21%), 5 (16%), 8 (13%), 12 (12%); frequent losses on 22q (16%), 9 (10%), 10 (8%). Recurrent partial imbalances comprised the minimal overlapping regions dim(10)(q25) and enh(12)(q13.3-q14.1). Unsupervised cluster analysis of genomic profiles detected two major subgroups (group I: complete gain of 7 and additional gains of 5, 8 or 12; group II: no major recurring imbalances, mainly losses). A comparison with low-grade gliomas (astrocytomas WHO grade II) showed chromosome 5 gain to be significantly more frequent in gangliogliomas. Interphase fluorescence in situ hybridization (FISH) identified the aberrations to be contained in a subpopulation of glial but not in neuronal cells. Two gangliogliomas and their anaplastic recurrences (WHO grade III) were analyzed. Losses of CDKN2A/B and DMBT1 or a gain/amplification of CDK4 found in the anaplastic tumors were already present in the respective gangliogliomas by array CGH and interphase FISH. In summary, genomic profiling in a large series of gangliogliomas could distinguish genetic subgroups even in this low-grade tumor.
Figures
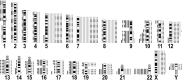

References
-
- Baudis M (2006) Online database and bioinformatics toolbox to support data mining in cancer cytogenetics. Biotechniques 40:269–270, 272. - PubMed
-
- Baudis M, Cleary ML (2001) Progenetix.net: an online repository for molecular cytogenetic aberration data. Bioinformatics 17:1228–1229. - PubMed
-
- Becker AJ, Lobach M, Klein H, Normann S, Nothen MM, Von Deimling A et al (2001) Mutational analysis of TSC1 and TSC2 genes in gangliogliomas. Neuropathol Appl Neurobiol 27:105–114. - PubMed
-
- Becker AJ, Klein H, Baden T, Aigner L, Normann S, Elger CE et al (2002) Mutational and expression analysis of the reelin pathway components CDK5 and doublecortin in gangliogliomas. Acta Neuropathol (Berl) 104:403–408. - PubMed
-
- Becker AJ, Blumcke I, Urbach H, Hans V, Majores M (2006) Molecular neuropathology of epilepsy‐associated glioneuronal malformations. J Neuropathol Exp Neurol 65:99–108. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

