par genes in Mycobacterium bovis and Mycobacterium smegmatis are arranged in an operon transcribed from "SigGC" promoters
- PMID: 18371202
- PMCID: PMC2346475
- DOI: 10.1186/1471-2180-8-51
par genes in Mycobacterium bovis and Mycobacterium smegmatis are arranged in an operon transcribed from "SigGC" promoters
Abstract
Background: The ParA/Soj and ParB/Spo0J proteins, and the cis-acting parS site, participate actively in chromosome segregation and cell cycle progression. Genes homologous to parA and parB, and two putative parS copies, have been identified in the Mycobacterium bovis BCG and Mycobacterium smegmatis chromosomes. As in Mycobacterium tuberculosis, the parA and parB genes in these two non-pathogenic mycobacteria are located near the chromosomal origin of replication. The present work focused on the determination of the transcriptional organisation of the ~6 Kb orf60K-parB region of M. bovis BCG and M. smegmatis by primer extension, transcriptional fusions to the green fluorescence protein (GFP) and quantitative RT-PCR.
Results: The parAB genes were arranged in an operon. However, we also found promoters upstream of each one of these genes. Seven putative promoter sequences were identified in the orf60K-parB region of M. bovis BCG, whilst four were identified in the homologous region of M. smegmatis, one upstream of each open reading frame (ORF).Real-time PCR assays showed that in M. smegmatis, mRNA-parA and mRNA-parB levels decreased between the exponential and stationary phases. In M. bovis BCG, mRNA-parA levels also decreased between the exponential and stationary phases. However, parB expression was higher than parA expression and remained almost unchanged along the growth curve.
Conclusion: The majority of the proposed promoter regions had features characteristic of Mycobacterium promoters previously denoted as Group D. The -10 hexamer of a strong E. coli sigma70-like promoter, located upstream of gidB of M. bovis BCG, overlapped with a putative parS sequence, suggesting that the transcription from this promoter might be regulated by the binding of ParB to parS.
Figures



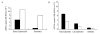
References
-
- Hiraga S. Chromosome and plasmid partition in Escherichia coli. Annu Rev Biochem. 1992;61:283–306. - PubMed
-
- Koonin EV. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol. 1993;229:1165–1174. - PubMed
-
- Mori H, Kondo A, Ohshima A, Ogura T, Hiraga S. Structure and function of the F ÿplasmid genes essential for partitioning. J Mol Biol. 1986;192:1–15. - PubMed
-
- Austin S, Abeles AL. Partitioning of unit copy miniplasmids to daughter cells. II. The partititon regions of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J Mol Biol. 1983;169:373–387. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

