Cophylogeny of the anther smut fungi and their caryophyllaceous hosts: prevalence of host shifts and importance of delimiting parasite species for inferring cospeciation
- PMID: 18371215
- PMCID: PMC2324105
- DOI: 10.1186/1471-2148-8-100
Cophylogeny of the anther smut fungi and their caryophyllaceous hosts: prevalence of host shifts and importance of delimiting parasite species for inferring cospeciation
Abstract
Background: Using phylogenetic approaches, the expectation that parallel cladogenesis should occur between parasites and hosts has been validated in some studies, but most others provided evidence for frequent host shifts. Here we examine the evolutionary history of the association between Microbotryum fungi that cause anther smut disease and their Caryophyllaceous hosts. We investigated the congruence between host and parasite phylogenies, inferred cospeciation events and host shifts, and assessed whether geography or plant ecology could have facilitated the putative host shifts identified. For cophylogeny analyses on microorganisms, parasite strains isolated from different host species are generally considered to represent independent evolutionary lineages, often without checking whether some strains actually belong to the same generalist species. Such an approach may mistake intraspecific nodes for speciation events and thus bias the results of cophylogeny analyses if generalist species are found on closely related hosts. A second aim of this study was therefore to evaluate the impact of species delimitation on the inferences of cospeciation.
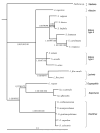
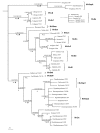
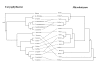
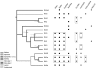
Results: We inferred a multiple gene phylogeny of anther smut strains from 21 host plants from several geographic origins, complementing a previous study on the delimitation of fungal species and their host specificities. We also inferred a multi-gene phylogeny of their host plants, and the two phylogenies were compared. A significant level of cospeciation was found when each host species was considered to harbour a specific parasite strain, i.e. when generalist parasite species were not recognized as such. This approach overestimated the frequency of cocladogenesis because individual parasite species capable of infecting multiple host species (i.e. generalists) were found on closely related hosts. When generalist parasite species were appropriately delimited and only a single representative of each species was retained, cospeciation events were not more frequent than expected under a random distribution, and many host shifts were inferred.Current geographic distributions of host species seemed to be of little relevance for understanding the putative historical host shifts, because most fungal species had overlapping geographic ranges. We did detect some ecological similarities, including shared pollinators and habitat types, between host species that were diseased by closely related anther smut species. Overall, genetic similarity underlying the host-parasite interactions appeared to have the most important influence on specialization and host-shifts: generalist multi-host parasite species were found on closely related plant species, and related species in the Microbotryum phylogeny were associated with members of the same host clade.
Conclusion: We showed here that Microbotryum species have evolved through frequent host shifts to moderately distant hosts, and we show further that accurate delimitation of parasite species is essential for interpreting cophylogeny studies.
Figures

Similar articles
-
Phylogenetic evidence of host-specific cryptic species in the anther smut fungus.Evolution. 2007 Jan;61(1):15-26. doi: 10.1111/j.1558-5646.2007.00002.x. Evolution. 2007. PMID: 17300424
-
Distribution of the anther-smut pathogen Microbotryum on species of the Caryophyllaceae.New Phytol. 2010 Jul;187(1):217-229. doi: 10.1111/j.1469-8137.2010.03268.x. Epub 2010 Apr 12. New Phytol. 2010. PMID: 20406409 Free PMC article.
-
Phylogenetic congruence of parasitic smut fungi (Anthracoidea, Anthracoideaceae) and their host plants (Carex, Cyperaceae): Cospeciation or host-shift speciation?Am J Bot. 2015 Jul;102(7):1108-14. doi: 10.3732/ajb.1500130. Epub 2015 Jul 14. Am J Bot. 2015. PMID: 26199367
-
Mating system of the anther smut fungus Microbotryum violaceum: selfing under heterothallism.Eukaryot Cell. 2008 May;7(5):765-75. doi: 10.1128/EC.00440-07. Epub 2008 Feb 15. Eukaryot Cell. 2008. PMID: 18281603 Free PMC article. Review. No abstract available.
-
Cophylogeny: insights from fish-parasite systems.Parassitologia. 2007 Sep;49(3):125-8. Parassitologia. 2007. PMID: 18410070 Review.
Cited by
-
Stripe smuts of grasses: one lineage or high levels of polyphyly?Persoonia. 2014 Dec;33:169-81. doi: 10.3767/003158514X685202. Epub 2014 Oct 6. Persoonia. 2014. PMID: 25737599 Free PMC article.
-
EUCALYPT: efficient tree reconciliation enumerator.Algorithms Mol Biol. 2015 Jan 23;10(1):3. doi: 10.1186/s13015-014-0031-3. eCollection 2015. Algorithms Mol Biol. 2015. PMID: 25648467 Free PMC article.
-
Sympatry and interference of divergent Microbotryum pathogen species.Ecol Evol. 2019 Apr 12;9(9):5457-5467. doi: 10.1002/ece3.5140. eCollection 2019 May. Ecol Evol. 2019. PMID: 31110694 Free PMC article.
-
Suillus indicus sp. nov. (Boletales, Basidiomycota), a new boletoid fungus from northwestern Himalayas, India.Mycology. 2015 Jan 2;6(1):35-41. doi: 10.1080/21501203.2014.988770. Epub 2014 Dec 11. Mycology. 2015. PMID: 26000197 Free PMC article.
-
Experimental hybridization and backcrossing reveal forces of reproductive isolation in Microbotryum.BMC Evol Biol. 2013 Oct 10;13:224. doi: 10.1186/1471-2148-13-224. BMC Evol Biol. 2013. PMID: 24112452 Free PMC article.
References
-
- Timms R, Read AF. What makes a specialist special? TREE. 1999;14(9):333–334. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

