LocateP: genome-scale subcellular-location predictor for bacterial proteins
- PMID: 18371216
- PMCID: PMC2375117
- DOI: 10.1186/1471-2105-9-173
LocateP: genome-scale subcellular-location predictor for bacterial proteins
Abstract
Background: In the past decades, various protein subcellular-location (SCL) predictors have been developed. Most of these predictors, like TMHMM 2.0, SignalP 3.0, PrediSi and Phobius, aim at the identification of one or a few SCLs, whereas others such as CELLO and Psortb.v.2.0 aim at a broader classification. Although these tools and pipelines can achieve a high precision in the accurate prediction of signal peptides and transmembrane helices, they have a much lower accuracy when other sequence characteristics are concerned. For instance, it proved notoriously difficult to identify the fate of proteins carrying a putative type I signal peptidase (SPIase) cleavage site, as many of those proteins are retained in the cell membrane as N-terminally anchored membrane proteins. Moreover, most of the SCL classifiers are based on the classification of the Swiss-Prot database and consequently inherited the inconsistency of that SCL classification. As accurate and detailed SCL prediction on a genome scale is highly desired by experimental researchers, we decided to construct a new SCL prediction pipeline: LocateP.
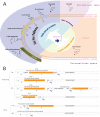
Results: LocateP combines many of the existing high-precision SCL identifiers with our own newly developed identifiers for specific SCLs. The LocateP pipeline was designed such that it mimics protein targeting and secretion processes. It distinguishes 7 different SCLs within Gram-positive bacteria: intracellular, multi-transmembrane, N-terminally membrane anchored, C-terminally membrane anchored, lipid-anchored, LPxTG-type cell-wall anchored, and secreted/released proteins. Moreover, it distinguishes pathways for Sec- or Tat-dependent secretion and alternative secretion of bacteriocin-like proteins. The pipeline was tested on data sets extracted from literature, including experimental proteomics studies. The tests showed that LocateP performs as well as, or even slightly better than other SCL predictors for some locations and outperforms current tools especially where the N-terminally anchored and the SPIase-cleaved secreted proteins are concerned. Overall, the accuracy of LocateP was always higher than 90%. LocateP was then used to predict the SCLs of all proteins encoded by completed Gram-positive bacterial genomes. The results are stored in the database LocateP-DB http://www.cmbi.ru.nl/locatep-db1.
Conclusion: LocateP is by far the most accurate and detailed protein SCL predictor for Gram-positive bacteria currently available.
Figures


References
-
- LocateP-DB http://www.cmbi.ru.nl/locatep-db
-
- Huang F, Parmryd I, Nilsson F, Persson AL, Pakrasi HB, Andersson B, Norling B. Proteomics of Synechocystis sp. strain PCC 6803: identification of plasma membrane proteins. Mol Cell Proteomics. 2002;1:956–966. - PubMed
-
- Molloy MP, Phadke ND, Maddock JR, Andrews PC. Two-dimensional electrophoresis and peptide mass fingerprinting of bacterial outer membrane proteins. Electrophoresis. 2001;22:1686–1696. - PubMed
-
- Molloy MP, Herbert BR, Slade MB, Rabilloud T, Nouwens AS, Williams KL, Gooley AA. Proteomic analysis of the Escherichia coli outer membrane. Eur J Biochem. 2000;267:2871–2881. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

