Brain gene expression profiles of Cln1 and Cln5 deficient mice unravels common molecular pathways underlying neuronal degeneration in NCL diseases
- PMID: 18371231
- PMCID: PMC2323392
- DOI: 10.1186/1471-2164-9-146
Brain gene expression profiles of Cln1 and Cln5 deficient mice unravels common molecular pathways underlying neuronal degeneration in NCL diseases
Abstract
Background: The neuronal ceroid lipofuscinoses (NCL) are a group of children's inherited neurodegenerative disorders, characterized by blindness, early dementia and pronounced cortical atrophy. The similar pathological and clinical profiles of the different forms of NCL suggest that common disease mechanisms may be involved. To explore the NCL-associated disease pathology and molecular pathways, we have previously produced targeted knock-out mice for Cln1 and Cln5. Both mouse-models replicate the NCL phenotype and neuropathology; the Cln1-/- model presents with early onset, severe neurodegenerative disease, whereas the Cln5-/- model produces a milder disease with a later onset.
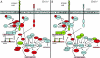
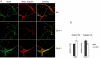
Results: Here we have performed quantitative gene expression profiling of the cortex from 1 and 4 month old Cln1-/- and Cln5-/- mice. Combined microarray datasets from both mouse models exposed a common affected pathway: genes regulating neuronal growth cone stabilization display similar aberrations in both models. We analyzed locus specific gene expression and showed regional clustering of Cln1 and three major genes of this pathway, further supporting a close functional relationship between the corresponding gene products; adenylate cyclase-associated protein 1 (Cap1), protein tyrosine phosphatase receptor type F (Ptprf) and protein tyrosine phosphatase 4a2 (Ptp4a2). The evidence from the gene expression data, indicating changes in the growth cone assembly, was substantiated by the immunofluorescence staining patterns of Cln1-/- and Cln5-/- cortical neurons. These primary neurons displayed abnormalities in cytoskeleton-associated proteins actin and beta-tubulin as well as abnormal intracellular distribution of growth cone associated proteins GAP-43, synapsin and Rab3.
Conclusion: Our data provide the first evidence for a common molecular pathogenesis behind neuronal degeneration in INCL and vLINCL. Since CLN1 and CLN5 code for proteins with distinct functional roles these data may have implications for other forms of NCLs as well.
Figures



Similar articles
-
Novel interactions of CLN5 support molecular networking between Neuronal Ceroid Lipofuscinosis proteins.BMC Cell Biol. 2009 Nov 26;10:83. doi: 10.1186/1471-2121-10-83. BMC Cell Biol. 2009. PMID: 19941651 Free PMC article.
-
Exacerbated neuronal ceroid lipofuscinosis phenotype in Cln1/5 double-knockout mice.Dis Model Mech. 2013 Mar;6(2):342-57. doi: 10.1242/dmm.010140. Epub 2012 Oct 12. Dis Model Mech. 2013. PMID: 23065637 Free PMC article.
-
Progress towards understanding disease mechanisms in small vertebrate models of neuronal ceroid lipofuscinosis.Biochim Biophys Acta. 2006 Oct;1762(10):873-89. doi: 10.1016/j.bbadis.2006.08.002. Epub 2006 Aug 10. Biochim Biophys Acta. 2006. PMID: 17023146 Review.
-
A mouse model for Finnish variant late infantile neuronal ceroid lipofuscinosis, CLN5, reveals neuropathology associated with early aging.Hum Mol Genet. 2004 Dec 1;13(23):2893-906. doi: 10.1093/hmg/ddh312. Epub 2004 Sep 30. Hum Mol Genet. 2004. PMID: 15459177
-
Cellular pathology and pathogenic aspects of neuronal ceroid lipofuscinoses.Adv Genet. 2001;45:35-68. doi: 10.1016/s0065-2660(01)45003-6. Adv Genet. 2001. PMID: 11332776 Review.
Cited by
-
Patterned expression of ion channel genes in mouse dorsal raphe nucleus determined with the Allen Mouse Brain Atlas.Brain Res. 2012 May 31;1457:1-12. doi: 10.1016/j.brainres.2012.03.066. Epub 2012 Apr 4. Brain Res. 2012. PMID: 22534482 Free PMC article.
-
Molecular elucidation of brain lipofuscin in aging and Neuronal Ceroid Lipofuscinosis.Res Sq [Preprint]. 2025 Mar 19:rs.3.rs-6010379. doi: 10.21203/rs.3.rs-6010379/v1. Res Sq. 2025. PMID: 40166029 Free PMC article. Preprint.
-
The Neuronal Ceroid Lipofuscinoses-Linked Loss of Function CLN5 and CLN8 Variants Disrupt Normal Lysosomal Function.Neuromolecular Med. 2019 Jun;21(2):160-169. doi: 10.1007/s12017-019-08529-7. Epub 2019 Mar 27. Neuromolecular Med. 2019. PMID: 30919163
-
Effects of heat shock protein 72 (Hsp72) on evolution of astrocyte activation following stroke in the mouse.Exp Neurol. 2012 Dec;238(2):284-96. doi: 10.1016/j.expneurol.2012.08.015. Epub 2012 Aug 20. Exp Neurol. 2012. PMID: 22940431 Free PMC article.
-
Novel interactions of CLN5 support molecular networking between Neuronal Ceroid Lipofuscinosis proteins.BMC Cell Biol. 2009 Nov 26;10:83. doi: 10.1186/1471-2121-10-83. BMC Cell Biol. 2009. PMID: 19941651 Free PMC article.
References
-
- Santavuori P. Neuronal ceroid-lipofuscinoses in childhood. Brain Dev. 1988;10:80–83. - PubMed
-
- Haltia M. The neuronal ceroid-lipofuscinoses: from past to present. Biochim Biophys Acta. 2006;1762:850–856. - PubMed
-
- Jalanko A, Vesa J, Manninen T, von Schantz C, Minye H, Fabritius AL, Salonen T, Rapola J, Gentile M, Kopra O, Peltonen L. Mice with Ppt1Deltaex4 mutation replicate the INCL phenotype and show an inflammation-associated loss of interneurons. Neurobiol Dis. 2005;18:226–241. doi: 10.1016/j.nbd.2004.08.013. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

