Methanogens with pseudomurein use diaminopimelate aminotransferase in lysine biosynthesis
- PMID: 18371309
- PMCID: PMC2424127
- DOI: 10.1016/j.febslet.2008.03.021
Methanogens with pseudomurein use diaminopimelate aminotransferase in lysine biosynthesis
Abstract
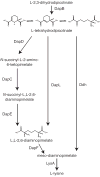
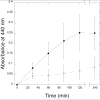
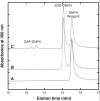
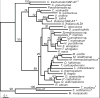
Methanothermobacter thermautotrophicus uses lysine for both protein synthesis and cross-linking pseudomurein in its cell wall. A diaminopimelate aminotransferase enzyme from this methanogen (MTH0052) converts tetrahydrodipicolinate to l,l-diaminopimelate, a lysine precursor. This gene complemented an Escherichia coli diaminopimelate auxotrophy, and the purified protein catalyzed the transamination of diaminopimelate to tetrahydrodipicolinate. Phylogenetic analysis indicated this gene was recruited from anaerobic Gram-positive bacteria. These results expand the family of diaminopimelate aminotransferases to a diverse set of plant, bacterial and archaeal homologs. In contrast marine methanogens from the Methanococcales, which lack pseudomurein, appear to use a different diaminopimelate pathway for lysine biosynthesis.
Figures

Similar articles
-
Methanococci use the diaminopimelate aminotransferase (DapL) pathway for lysine biosynthesis.J Bacteriol. 2010 Jul;192(13):3304-10. doi: 10.1128/JB.00172-10. Epub 2010 Apr 23. J Bacteriol. 2010. PMID: 20418392 Free PMC article.
-
Biochemical and phylogenetic characterization of a novel diaminopimelate biosynthesis pathway in prokaryotes identifies a diverged form of LL-diaminopimelate aminotransferase.J Bacteriol. 2008 May;190(9):3256-63. doi: 10.1128/JB.01381-07. Epub 2008 Feb 29. J Bacteriol. 2008. PMID: 18310350 Free PMC article.
-
The redundant aminotransferases in lysine and arginine synthesis and the extent of aminotransferase redundancy in Escherichia coli.Mol Microbiol. 2014 Nov;94(4):843-56. doi: 10.1111/mmi.12801. Epub 2014 Oct 12. Mol Microbiol. 2014. PMID: 25243376
-
Two major archaeal pseudomurein endoisopeptidases: PeiW and PeiP.Archaea. 2010 Nov 11;2010:480492. doi: 10.1155/2010/480492. Archaea. 2010. PMID: 21113291 Free PMC article. Review.
-
Enzymology of bacterial lysine biosynthesis.Adv Enzymol Relat Areas Mol Biol. 1998;72:279-324. doi: 10.1002/9780470123188.ch8. Adv Enzymol Relat Areas Mol Biol. 1998. PMID: 9559056 Review.
Cited by
-
Methanococci use the diaminopimelate aminotransferase (DapL) pathway for lysine biosynthesis.J Bacteriol. 2010 Jul;192(13):3304-10. doi: 10.1128/JB.00172-10. Epub 2010 Apr 23. J Bacteriol. 2010. PMID: 20418392 Free PMC article.
-
Outer and inner membrane proteins compose an arginine-agmatine exchange system in Chlamydophila pneumoniae.J Bacteriol. 2008 Nov;190(22):7431-40. doi: 10.1128/JB.00652-08. Epub 2008 Sep 12. J Bacteriol. 2008. PMID: 18790867 Free PMC article.
-
Identification and characterization of archaeal pseudomurein biosynthesis genes through pangenomics.mSystems. 2025 Mar 18;10(3):e0140124. doi: 10.1128/msystems.01401-24. Epub 2025 Feb 12. mSystems. 2025. PMID: 39936904 Free PMC article.
-
Cohesion group approach for evolutionary analysis of aspartokinase, an enzyme that feeds a branched network of many biochemical pathways.Microbiol Mol Biol Rev. 2009 Dec;73(4):594-651. doi: 10.1128/MMBR.00024-09. Microbiol Mol Biol Rev. 2009. PMID: 19946135 Free PMC article. Review.
References
-
- Scapin G, Blanchard JS. Enzymology of bacterial lysine biosynthesis. Adv Enzymol Relat Areas Mol Biol. 1998;72:279–324. - PubMed
-
- Xu H, Andi B, Qian J, West AH, Cook PF. The α-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem Biophys. 2006;46:43–64. - PubMed
-
- Ekiel I, Jarrell KF, Sprott GD. Amino acid biosynthesis and sodium-dependent transport in Methanococcus voltae, as revealed by 13C NMR. Eur J Biochem. 1985;149:437–444. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

