Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors
- PMID: 18371366
- PMCID: PMC2234224
- DOI: 10.1016/j.stem.2007.05.019
Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors
Abstract
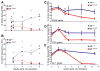
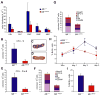
The Mixed Lineage Leukemia (MLL) gene is essential for embryonic hematopoietic stem cell (HSC) development, but its role during adult hematopoiesis is unknown. Using an inducible knockout model, we demonstrate that Mll is essential for the maintenance of adult HSCs and progenitors, with fatal bone marrow failure occurring within 3 weeks of Mll deletion. Mll-deficient cells are selectively lost from mixed bone marrow chimeras, demonstrating their failure to self-renew even in an intact bone marrow environment. Surprisingly, HSCs lacking Mll exhibit ectopic cell-cycle entry, resulting in the depletion of quiescent HSCs. In contrast, Mll deletion in myelo-erythroid progenitors results in reduced proliferation and reduced response to cytokine-induced cell-cycle entry. Committed lymphoid and myeloid cells no longer require Mll, defining the early multipotent stages of hematopoiesis as Mll dependent. These studies demonstrate that Mll plays selective and independent roles within the hematopoietic system, maintaining quiescence in HSCs and promoting proliferation in progenitors.
Figures

Similar articles
-
Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal.Cell Stem Cell. 2007 Sep 13;1(3):338-45. doi: 10.1016/j.stem.2007.07.002. Cell Stem Cell. 2007. PMID: 18371367
-
Stress hematopoiesis reveals abnormal control of self-renewal, lineage bias, and myeloid differentiation in Mll partial tandem duplication (Mll-PTD) hematopoietic stem/progenitor cells.Blood. 2012 Aug 2;120(5):1118-29. doi: 10.1182/blood-2012-02-412379. Epub 2012 Jun 26. Blood. 2012. PMID: 22740449 Free PMC article.
-
MLL-ENL cooperates with SCF to transform primary avian multipotent cells.EMBO J. 2002 Aug 15;21(16):4297-306. doi: 10.1093/emboj/cdf429. EMBO J. 2002. PMID: 12169632 Free PMC article.
-
Leukemogenesis via aberrant self-renewal by the MLL/AEP-mediated transcriptional activation system.Cancer Sci. 2021 Oct;112(10):3935-3944. doi: 10.1111/cas.15054. Epub 2021 Aug 2. Cancer Sci. 2021. PMID: 34251718 Free PMC article. Review.
-
Exploring the contribution of Zfp521/ZNF521 on primary hematopoietic stem/progenitor cells and leukemia progression.Cell Tissue Res. 2024 Dec;398(3):161-173. doi: 10.1007/s00441-024-03926-2. Epub 2024 Oct 22. Cell Tissue Res. 2024. PMID: 39436449 Free PMC article. Review.
Cited by
-
Mixed lineage leukemia 5 (MLL5) protein regulates cell cycle progression and E2F1-responsive gene expression via association with host cell factor-1 (HCF-1).J Biol Chem. 2013 Jun 14;288(24):17532-43. doi: 10.1074/jbc.M112.439729. Epub 2013 Apr 29. J Biol Chem. 2013. PMID: 23629655 Free PMC article.
-
Dual function of histone H3 lysine 36 methyltransferase ASH1 in regulation of Hox gene expression.PLoS One. 2011;6(11):e28171. doi: 10.1371/journal.pone.0028171. Epub 2011 Nov 28. PLoS One. 2011. PMID: 22140534 Free PMC article.
-
Runx1 loss minimally impacts long-term hematopoietic stem cells.PLoS One. 2011;6(12):e28430. doi: 10.1371/journal.pone.0028430. Epub 2011 Dec 1. PLoS One. 2011. PMID: 22145044 Free PMC article.
-
BRPF1 is essential for development of fetal hematopoietic stem cells.J Clin Invest. 2016 Sep 1;126(9):3247-62. doi: 10.1172/JCI80711. Epub 2016 Aug 8. J Clin Invest. 2016. PMID: 27500495 Free PMC article.
-
Hematopoietic stem and progenitor cell proliferation and differentiation requires the trithorax protein Ash2l.Sci Rep. 2019 Jun 4;9(1):8262. doi: 10.1038/s41598-019-44720-3. Sci Rep. 2019. PMID: 31164666 Free PMC article.
References
-
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. - PubMed
-
- Ayton P, Sneddon SF, Palmer DB, Rosewell IR, Owen MJ, Young B, Presley R, Subramanian V. Truncation of the Mll gene in exon 5 by gene targeting leads to early preimplantation lethality of homozygous embryos. Genesis. 2001;30:201–212. - PubMed
-
- Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

