Epigenetic regulation of heterochromatic DNA stability
- PMID: 18372168
- PMCID: PMC2814359
- DOI: 10.1016/j.gde.2008.01.021
Epigenetic regulation of heterochromatic DNA stability
Abstract
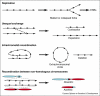
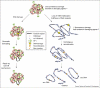
In this review we summarize recent studies that demonstrate the importance of epigenetic mechanisms for maintaining genome integrity, specifically with respect to repeated DNAs within heterochromatin. Potential problems that arise during replication, recombination, and repair of repeated sequences are counteracted by post-translational histone modifications and associated proteins, including the cohesins. These factors appear to ensure repeat stability by multiple mechanisms: suppressing homologous recombination, controlling the three-dimensional organization of damaged repeats to reduce the probability of aberrant recombination, and promoting the use of less problematic repair pathways. The presence of such systems may facilitate repeat and chromosome evolution, and their failure can lead to genome instability, chromosome rearrangements, and the onset of pathogenesis.
Figures
References
-
- John B. The biology of heterochromatin. In: Verma RS, editor. Heterochromatin: Molecular Structural Aspects. Cambridge University Press; 1988. pp. 1–147.
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Heitz E. Das heterochromatin der moose. Jahrb Wiss Bot. 1928;69:762–818.
-
- Karpen GH, Le MH, Le H. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science. 1996;273:118–122. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

