FISH mapping of de novo apparently balanced chromosome rearrangements identifies characteristics associated with phenotypic abnormality
- PMID: 18374296
- PMCID: PMC2491339
- DOI: 10.1016/j.ajhg.2008.02.007
FISH mapping of de novo apparently balanced chromosome rearrangements identifies characteristics associated with phenotypic abnormality
Erratum in
- Am J Hum Genet. 2008 Apr;82(4):1019
Abstract
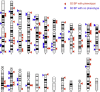
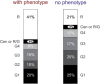
We report fluorescence in situ hybridization (FISH) mapping of 152, mostly de novo, apparently balanced chromosomal rearrangement (ABCR) breakpoints in 76 individuals, 30 of whom had no obvious phenotypic abnormality (control group) and 46 of whom had an associated disease (case group). The aim of this study was to identify breakpoint characteristics that could discriminate between these groups and which might be of predictive value in de novo ABCR (DN-ABCR) cases detected antenatally. We found no difference in the proportion of breakpoints that interrupted a gene, although in three cases, direct interruption or deletion of known autosomal-dominant or X-linked recessive Mendelian disease genes was diagnostic. The only significant predictor of phenotypic abnormality in the group as a whole was the localization of one or both breakpoints to an R-positive (G-negative) band with estimated predictive values of 0.69 (95% CL 0.54-0.81) and 0.90 (95% CL 0.60-0.98), respectively. R-positive bands are known to contain more genes and have a higher guanine-cytosine (GC) content than do G-positive (R-negative) bands; however, whether a gene was interrupted by the breakpoint or the GC content in the 200 kB around the breakpoint had no discriminant ability. Our results suggest that the large-scale genomic context of the breakpoint has prognostic utility and that the pathological mechanism of mapping to an R-band cannot be accounted for by direct gene inactivation.
Figures
Similar articles
-
Breakpoint mapping by next generation sequencing reveals causative gene disruption in patients carrying apparently balanced chromosome rearrangements with intellectual deficiency and/or congenital malformations.J Med Genet. 2013 Mar;50(3):144-50. doi: 10.1136/jmedgenet-2012-101351. Epub 2013 Jan 12. J Med Genet. 2013. PMID: 23315544
-
The clinical impact of chromosomal rearrangements with breakpoints upstream of the SOX9 gene: two novel de novo balanced translocations associated with acampomelic campomelic dysplasia.BMC Med Genet. 2013 May 7;14:50. doi: 10.1186/1471-2350-14-50. BMC Med Genet. 2013. PMID: 23648064 Free PMC article.
-
Complex nature of apparently balanced chromosomal rearrangements in patients with autism spectrum disorder.Mol Autism. 2015 Mar 25;6:19. doi: 10.1186/s13229-015-0015-2. eCollection 2015. Mol Autism. 2015. PMID: 25844147 Free PMC article.
-
Prenatal diagnosis of a fetus with two balanced de novo chromosome rearrangements.Am J Med Genet. 1996 Dec 11;66(2):197-9. doi: 10.1002/(SICI)1096-8628(19961211)66:2<197::AID-AJMG14>3.0.CO;2-O. Am J Med Genet. 1996. PMID: 8958330 Review.
-
Prenatal diagnosis of de novo X;autosome translocations.Clin Genet. 2004 May;65(5):423-8. doi: 10.1111/j.0009-9163.2004.00255.x. Clin Genet. 2004. PMID: 15099352 Review.
Cited by
-
Severe Progressive Autism Associated with Two de novo Changes: A 2.6-Mb 2q31.1 Deletion and a Balanced t(14;21)(q21.1;p11.2) Translocation with Long-Range Epigenetic Silencing of LRFN5 Expression.Mol Syndromol. 2010 Feb;1(1):46-57. doi: 10.1159/000280290. Epub 2010 Feb 12. Mol Syndromol. 2010. PMID: 20648246 Free PMC article.
-
NR2F1 deletion in a patient with a de novo paracentric inversion, inv(5)(q15q33.2), and syndromic deafness.Am J Med Genet A. 2009 May;149A(5):931-8. doi: 10.1002/ajmg.a.32764. Am J Med Genet A. 2009. PMID: 19353646 Free PMC article.
-
Translocations disrupting PHF21A in the Potocki-Shaffer-syndrome region are associated with intellectual disability and craniofacial anomalies.Am J Hum Genet. 2012 Jul 13;91(1):56-72. doi: 10.1016/j.ajhg.2012.05.005. Epub 2012 Jul 5. Am J Hum Genet. 2012. PMID: 22770980 Free PMC article.
-
Chromosomal Translocations Detection in Cancer Cells Using Chromosomal Conformation Capture Data.Genes (Basel). 2022 Jun 29;13(7):1170. doi: 10.3390/genes13071170. Genes (Basel). 2022. PMID: 35885953 Free PMC article.
-
Detection of chromosomal breakpoints in patients with developmental delay and speech disorders.PLoS One. 2014 Mar 6;9(6):e90852. doi: 10.1371/journal.pone.0090852. eCollection 2014. PLoS One. 2014. PMID: 24603971 Free PMC article.
References
-
- Ray P.N., Belfall B., Duff C., Logan C., Kean V., Thompson M.W., Sylvester J.E., Gorski J.L., Schmickel R.D., Worton R.G. Cloning of the breakpoint of an X;21 translocation associated with Duchenne muscular dystrophy. Nature. 1985;318:672–675. - PubMed
-
- Mercer J.F., Livingston J., Hall B., Paynter J.A., Begy C., Chandrasekharappa S., Lockhart P., Grimes A., Bhave M., Siemieniak D. Isolation of a partial candidate gene for Menkes disease by positional cloning. Nat. Genet. 1993;3:20–25. - PubMed
-
- Nishimura D.Y., Swiderski R.E., Alward W.L., Searby C.C., Patil S.R., Bennet S.R., Kanis A.B., Gastier J.M., Stone E.M., Sheffield V.C. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat. Genet. 1998;19:140–147. - PubMed
-
- Kurotaki N., Imaizumi K., Harada N., Masuno M., Kondoh T., Nagai T., Ohashi H., Naritomi K., Tsukahara M., Makita Y. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 2002;30:365–366. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

