Brain regeneration from pluripotent stem cells in planarian
- PMID: 18375378
- PMCID: PMC2610179
- DOI: 10.1098/rstb.2008.2260
Brain regeneration from pluripotent stem cells in planarian
Abstract
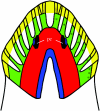
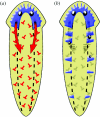
How can planarians regenerate their brain? Recently we have identified many genes critical for this process. Brain regeneration can be divided into five steps: (1) anterior blastema formation, (2) brain rudiment formation, (3) pattern formation, (4) neural network formation, and (5) functional recovery. Here we will describe the structure and process of regeneration of the planarian brain in the first part, and then introduce genes involved in brain regeneration in the second part. Especially, we will speculate about molecular events during the early steps of brain regeneration in this review. The finding providing the greatest insight thus far is the discovery of the nou-darake (ndk; 'brains everywhere' in Japanese) gene, since brain neurons are formed throughout the entire body as a result of loss of function of the ndk gene. This finding provides a clue for elucidating the molecular and cellular mechanisms underlying brain regeneration. Here we describe the molecular action of the nou-darake gene and propose a new model to explain brain regeneration and restriction in the head region of the planarians.
Figures



References
-
- Agata K. Regeneration and gene regulation in planarians. Curr. Opin. Genet. Dev. 2003;13:492–496. doi:10.1016/j.gde.2003.08.009 - DOI - PubMed
-
- Agata K, Watanabe K. Molecular and cellular aspects of planarian regeneration. Semin. Cell Dev. Biol. 1999;10:377–383. doi:10.1006/scdb.1999.0324 - DOI - PubMed
-
- Agata K, Soejima Y, Kato K, Kobayashi C, Umesono Y, Watanabe K. Structure of the planarian central nervous system (CNS) revealed by neuronal cell markers. Zool. Sci. 1998;15:433–440. doi:10.2108/zsj.15.433 - DOI - PubMed
-
- Agata K, Tanaka T, Kobayashi C, Kato K, Saitoh Y. Intercalary regeneration in planarians. Dev. Dyn. 2003;226:308–316. doi:10.1002/dvdy.10249 - DOI - PubMed
-
- Agata K, Nakajima E, Funayama N, Shibata N, Saito Y, Umesono Y. Two different evolutionary origins of stem cell systems and their molecular basis. Semin. Cell Dev. Biol. 2006;17:503–509. doi:10.1016/j.semcdb.2006.05.004 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
