Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses
- PMID: 18375600
- PMCID: PMC2330312
- DOI: 10.1104/pp.108.115733
Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses
Abstract
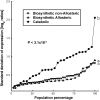
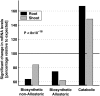
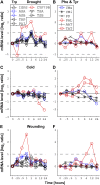
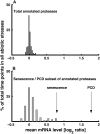
Using a bioinformatics analysis of public Arabidopsis (Arabidopsis thaliana) microarray data, we propose here a novel regulatory program, combining transcriptional and posttranslational controls, which participate in modulating fluxes of amino acid metabolism in response to abiotic stresses. The program includes the following two components: (1) the terminal enzyme of the module, responsible for the first catabolic step of the amino acid, whose level is stimulated or repressed in response to stress cues, just-in-time when the cues arrive, principally via transcriptional regulation of its gene; and (2) the initiator enzyme of the module, whose activity is principally modulated via posttranslational allosteric feedback inhibition in response to changes in the level of the amino acid, just-in-case when it occurs in response to alteration in its catabolism or sequestration into different intracellular compartments. Our proposed regulatory program is based on bioinformatics dissection of the response of all biosynthetic and catabolic genes of seven different pathways, involved in the metabolism of 11 amino acids, to eight different abiotic stresses, as judged from modulations of their mRNA levels. Our results imply that the transcription of the catabolic genes is principally more sensitive than that of the biosynthetic genes to fluctuations in stress-associated signals. Notably, the only exception to this program is the metabolic pathway of Pro, an amino acid that distinctively accumulates to significantly high levels under abiotic stresses. Examples of the biological significance of our proposed regulatory program are discussed.
Figures




References
-
- Amir R, Hacham Y, Galili G (2002) Cystathionine gamma-synthase and threonine synthase operate in concert to regulate carbon flow towards methionine in plants. Trends Plant Sci 7 153–156 - PubMed
-
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, Leaver CJ (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42 567–585 - PubMed
-
- Domany E (2005) Analysis of DNA-chip and antigen-chip data: studies of cancer, stem cells and autoimmune diseases. Comput Phys Commun 169 183–187
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

