Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues
- PMID: 18375788
- PMCID: PMC2327352
- DOI: 10.1261/rna.939908
Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues
Abstract
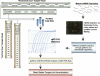
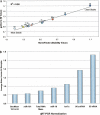
Proper normalization is a critical but often an underappreciated aspect of quantitative gene expression analysis. This study describes the identification and characterization of appropriate reference RNA targets for the normalization of microRNA (miRNA) quantitative RT-PCR data. miRNA microarray data from dozens of normal and disease human tissues revealed ubiquitous and stably expressed normalization candidates for evaluation by qRT-PCR. miR-191 and miR-103, among others, were found to be highly consistent in their expression across 13 normal tissues and five pair of distinct tumor/normal adjacent tissues. These miRNAs were statistically superior to the most commonly used reference RNAs used in miRNA qRT-PCR experiments, such as 5S rRNA, U6 snRNA, or total RNA. The most stable normalizers were also highly conserved across flash-frozen and formalin-fixed paraffin-embedded lung cancer tumor/NAT sample sets, resulting in the confirmation of one well-documented oncomir (let-7a), as well as the identification of novel oncomirs. These findings constitute the first report describing the rigorous normalization of miRNA qRT-PCR data and have important implications for proper experimental design and accurate data interpretation.
Figures

References
-
- Ambros, V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
-
- Andersen, C.L., Jensen, J.L., Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. - PubMed
-
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Bas, A., Forsberg, G., Hammarström, S., Hammarström, M.L. Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand. J. Immunol. 2004;6:566–573. - PubMed
-
- Calin, G.A., Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006a;6:857–866. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
