The signatures of autozygosity among patients with colorectal cancer
- PMID: 18375840
- PMCID: PMC4383032
- DOI: 10.1158/0008-5472.CAN-07-5250
The signatures of autozygosity among patients with colorectal cancer
Abstract
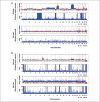
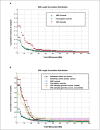
Previous studies have shown that among populations with a high rate of consanguinity, there is a significant increase in the prevalence of cancer. Single nucleotide polymorphism (SNP) array data (Affymetrix, 50K XbaI) analysis revealed long regions of homozygosity in genomic DNAs taken from tumor and matched normal tissues of colorectal cancer (CRC) patients. The presence of these regions in the genome may indicate levels of consanguinity in the individual's family lineage. We refer to these autozygous regions as identity-by-descent (IBD) segments. In this study, we compared IBD segments in 74 mostly Caucasian CRC patients (mean age of 66 years) to two control data sets: (a) 146 Caucasian individuals (mean age of 80 years) who participated in an age-related macular degeneration (AMD) study and (b) 118 cancer-free Caucasian individuals from the Framingham Heart Study (mean age of 67 years). Our results show that the percentage of CRC patients with IBD segments (>or=4 Mb length and 50 SNPs probed) in the genome is at least twice as high as the AMD or Framingham control groups. Also, the average length of these IBD regions in the CRC patients is more than twice the length of the two control data sets. Compared with control groups, IBD segments are found to be more common among individuals of Jewish background. We believe that these IBD segments within CRC patients are likely to harbor important CRC-related genes with low-penetrance SNPs and/or mutations, and, indeed, two recently identified CRC predisposition SNPs in the 8q24 region were confirmed to be homozygous in one particular patient carrying an IBD segment covering the region.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures

References
-
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. - PubMed
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. - PubMed
-
- Ahmed FE. Colon cancer: prevalence, screening, gene expression and mutation, and risk factors and assessment. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2003;21:65–131. - PubMed
-
- de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4:769–80. - PubMed
-
- Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

