Discrete state model and accurate estimation of loop entropy of RNA secondary structures
- PMID: 18376982
- PMCID: PMC2494904
- DOI: 10.1063/1.2895050
Discrete state model and accurate estimation of loop entropy of RNA secondary structures
Abstract
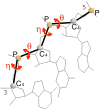
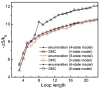
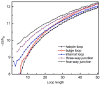
Conformational entropy makes important contribution to the stability and folding of RNA molecule, but it is challenging to either measure or compute conformational entropy associated with long loops. We develop optimized discrete k-state models of RNA backbone based on known RNA structures for computing entropy of loops, which are modeled as self-avoiding walks. To estimate entropy of hairpin, bulge, internal loop, and multibranch loop of long length (up to 50), we develop an efficient sampling method based on the sequential Monte Carlo principle. Our method considers excluded volume effect. It is general and can be applied to calculating entropy of loops with longer length and arbitrary complexity. For loops of short length, our results are in good agreement with a recent theoretical model and experimental measurement. For long loops, our estimated entropy of hairpin loops is in excellent agreement with the Jacobson-Stockmayer extrapolation model. However, for bulge loops and more complex secondary structures such as internal and multibranch loops, we find that the Jacobson-Stockmayer extrapolation model has large errors. Based on estimated entropy, we have developed empirical formulae for accurate calculation of entropy of long loops in different secondary structures. Our study on the effect of asymmetric size of loops suggest that loop entropy of internal loops is largely determined by the total loop length, and is only marginally affected by the asymmetric size of the two loops. Our finding suggests that the significant asymmetric effects of loop length in internal loops measured by experiments are likely to be partially enthalpic. Our method can be applied to develop improved energy parameters important for studying RNA stability and folding, and for predicting RNA secondary and tertiary structures. The discrete model and the program used to calculate loop entropy can be downloaded at http://gila.bioengr.uic.edu/resources/RNA.html.
Figures








Similar articles
-
A two-length-scale polymer theory for RNA loop free energies and helix stacking.RNA. 2010 Jul;16(7):1350-5. doi: 10.1261/rna.1831710. Epub 2010 May 26. RNA. 2010. PMID: 20504955 Free PMC article.
-
Exact calculation of loop formation probability identifies folding motifs in RNA secondary structures.RNA. 2016 Dec;22(12):1808-1818. doi: 10.1261/rna.053694.115. Epub 2016 Oct 19. RNA. 2016. PMID: 27852924 Free PMC article.
-
Role of small loops in DNA melting.Biopolymers. 2001 Apr 5;58(4):374-89. doi: 10.1002/1097-0282(20010405)58:4<374::AID-BIP1014>3.0.CO;2-P. Biopolymers. 2001. PMID: 11180051
-
RNA folding: conformational statistics, folding kinetics, and ion electrostatics.Annu Rev Biophys. 2008;37:197-214. doi: 10.1146/annurev.biophys.37.032807.125957. Annu Rev Biophys. 2008. PMID: 18573079 Free PMC article. Review.
-
RNA structure at high resolution.FASEB J. 1995 Aug;9(11):1023-33. doi: 10.1096/fasebj.9.11.7544309. FASEB J. 1995. PMID: 7544309 Review.
Cited by
-
Equilibrium sampling in biomolecular simulations.Annu Rev Biophys. 2011;40:41-62. doi: 10.1146/annurev-biophys-042910-155255. Annu Rev Biophys. 2011. PMID: 21370970 Free PMC article. Review.
-
Loop Entropy Assists Tertiary Order: Loopy Stabilization of Stacking Motifs.Entropy (Basel). 2011 Nov 24;13(11):1958-1966. doi: 10.3390/e13111958. Entropy (Basel). 2011. PMID: 26379416 Free PMC article.
-
Physics-based RNA structure prediction.Biophys Rep. 2015;1:2-13. doi: 10.1007/s41048-015-0001-4. Epub 2015 Jul 9. Biophys Rep. 2015. PMID: 26942214 Free PMC article.
-
Fast exploration of an optimal path on the multidimensional free energy surface.PLoS One. 2017 May 18;12(5):e0177740. doi: 10.1371/journal.pone.0177740. eCollection 2017. PLoS One. 2017. PMID: 28542475 Free PMC article.
-
A conditional random fields method for RNA sequence-structure relationship modeling and conformation sampling.Bioinformatics. 2011 Jul 1;27(13):i102-10. doi: 10.1093/bioinformatics/btr232. Bioinformatics. 2011. PMID: 21685058 Free PMC article.
References
-
- Mathews DH, Sabina J, Zuker M, Turner DH. J Mol Biol. 1999;288:911. - PubMed
-
- Serra MJ, Turner DH. Methods Enzymol. 1995;259:242. - PubMed
-
- Xia T, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Biochemistry. 1998;37:14719. - PubMed
-
- Diamond JM, Turner DH, Mathews DH. Biochemistry. 2001;40:6971. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

