The unique genomic properties of sex-biased genes: insights from avian microarray data
- PMID: 18377635
- PMCID: PMC2294128
- DOI: 10.1186/1471-2164-9-148
The unique genomic properties of sex-biased genes: insights from avian microarray data
Abstract
Background: In order to develop a framework for the analysis of sex-biased genes, we present a characterization of microarray data comparing male and female gene expression in 18 day chicken embryos for brain, gonad, and heart tissue.
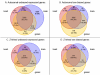
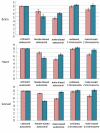
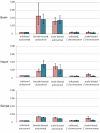
Results: From the 15982 significantly expressed coding regions that have been assigned to either the autosomes or the Z chromosome (12979 in brain, 13301 in gonad, and 12372 in heart), roughly 18% were significantly sex-biased in any one tissue, though only 4 gene targets were biased in all tissues. The gonad was the most sex-biased tissue, followed by the brain. Sex-biased autosomal genes tended to be expressed at lower levels and in fewer tissues than unbiased gene targets, and autosomal somatic sex-biased genes had more expression noise than similar unbiased genes. Sex-biased genes linked to the Z-chromosome showed reduced expression in females, but not in males, when compared to unbiased Z-linked genes, and sex-biased Z-linked genes were also expressed in fewer tissues than unbiased Z coding regions. Third position GC content, and codon usage bias showed some sex-biased effects, primarily for autosomal genes expressed in the gonad. Finally, there were several over-represented Gene Ontology terms in the sex-biased gene sets.
Conclusion: On the whole, this analysis suggests that sex-biased genes have unique genomic and organismal properties that delineate them from genes that are expressed equally in males and females.
Figures

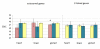

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

