Anaerobic consumers of monosaccharides in a moderately acidic fen
- PMID: 18378662
- PMCID: PMC2394944
- DOI: 10.1128/AEM.00193-08
Anaerobic consumers of monosaccharides in a moderately acidic fen
Abstract
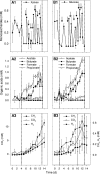
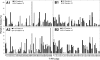
16S rRNA-based stable isotope probing identified active xylose- and glucose-fermenting Bacteria and active Archaea, including methanogens, in anoxic slurries of material obtained from a moderately acidic, CH(4)-emitting fen. Xylose and glucose were converted to fatty acids, CO(2), H(2), and CH(4) under moderately acidic, anoxic conditions, indicating that the fen harbors moderately acid-tolerant xylose- and glucose-using fermenters, as well as moderately acid-tolerant methanogens. Organisms of the families Acidaminococcaceae, Aeromonadaceae, Clostridiaceae, Enterobacteriaceae, and Pseudomonadaceae and the order Actinomycetales, including hitherto unknown organisms, utilized xylose- or glucose-derived carbon, suggesting that highly diverse facultative aerobes and obligate anaerobes contribute to the flow of carbon in the fen under anoxic conditions. Uncultured Euryarchaeota (i.e., Methanosarcinaceae and Methanobacteriaceae) and Crenarchaeota species were identified by 16S rRNA analysis of anoxic slurries, demonstrating that the acidic fen harbors novel methanogens and Crenarchaeota organisms capable of anaerobiosis. Fermentation-derived molecules are conceived to be the primary drivers of methanogenesis when electron acceptors other than CO(2) are absent, and the collective findings of this study indicate that fen soils harbor diverse, acid-tolerant, and novel xylose-utilizing as well as glucose-utilizing facultative aerobes and obligate anaerobes that form trophic links to novel moderately acid-tolerant methanogens.
Figures



References
-
- Ahmed, Z., H. Banu, M. M. Rahman, F. Akhter, and M. S. Haque. 2001. Microbial activity on the degradation of lignocellulosic polysaccharides. Int. J. Biol. Sci. 1:993-997.
-
- Aselmann, I., and P. J. Crutzen. 1989. Global distribution of natural fresh water wetlands and rice paddies, their net primary productivity, seasonality and possible methane emission. J. Atmos. Chem. 8:307-359.
-
- Boone, D. R., W. B. Whitman, and P. Rouvière. 1993. Diversity and taxonomy of methanogens, p. 35-80. In J. G. Ferry (ed.), Methanogenesis. Chapman & Hall, New York, NY.
-
- Brenner, D. J., N. R. Krieg, J. T. Staley, G. M. Garrity, et al. (ed.). 2005. Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, the Proteobacteria. Part B, the Gammaproteobacteria. Springer, New York, NY.
-
- Brenner, D. J., N. R. Krieg, J. T. Staley, G. M. Garrity, et al. (ed.). 2005. Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, the Proteobacteria. Part C, the Alpha-, Beta-, Delta-, and Epsilonproteobacteria. Springer, New York, NY.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

