Functional diversity and electron donor dependence of microbial populations capable of U(VI) reduction in radionuclide-contaminated subsurface sediments
- PMID: 18378664
- PMCID: PMC2394950
- DOI: 10.1128/AEM.02881-07
Functional diversity and electron donor dependence of microbial populations capable of U(VI) reduction in radionuclide-contaminated subsurface sediments
Abstract
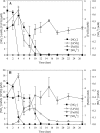
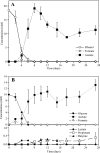
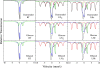
In order to elucidate the potential mechanisms of U(VI) reduction for the optimization of bioremediation strategies, the structure-function relationships of microbial communities were investigated in microcosms of subsurface materials cocontaminated with radionuclides and nitrate. A polyphasic approach was used to assess the functional diversity of microbial populations likely to catalyze electron flow under conditions proposed for in situ uranium bioremediation. The addition of ethanol and glucose as supplemental electron donors stimulated microbial nitrate and Fe(III) reduction as the predominant terminal electron-accepting processes (TEAPs). U(VI), Fe(III), and sulfate reduction overlapped in the glucose treatment, whereas U(VI) reduction was concurrent with sulfate reduction but preceded Fe(III) reduction in the ethanol treatments. Phyllosilicate clays were shown to be the major source of Fe(III) for microbial respiration by using variable-temperature Mössbauer spectroscopy. Nitrate- and Fe(III)-reducing bacteria (FeRB) were abundant throughout the shifts in TEAPs observed in biostimulated microcosms and were affiliated with the genera Geobacter, Tolumonas, Clostridium, Arthrobacter, Dechloromonas, and Pseudomonas. Up to two orders of magnitude higher counts of FeRB and enhanced U(VI) removal were observed in ethanol-amended treatments compared to the results in glucose-amended treatments. Quantification of citrate synthase (gltA) levels demonstrated a stimulation of Geobacteraceae activity during metal reduction in carbon-amended microcosms, with the highest expression observed in the glucose treatment. Phylogenetic analysis indicated that the active FeRB share high sequence identity with Geobacteraceae members cultivated from contaminated subsurface environments. Our results show that the functional diversity of populations capable of U(VI) reduction is dependent upon the choice of electron donor.
Figures

References
-
- Abdelouas, A., W. Lutze, and H. E. Nuttall. 1999. Uranium contamination in the subsurface; characterization and remediation. Rev. Mineral. Geochem. 38:433-473.
-
- Akob, D. M., H. J. Mills, and J. E. Kostka. 2007. Metabolically active microbial communities in uranium-contaminated subsurface sediments. FEMS Microbiol. Ecol. 59:95-107. - PubMed
-
- Alef, K. 1991. Methodenhandbuch Bodenmikrobiologie: Aktivitaten, Biomasse, Differenzierung, p. 44-49. Ecomed, Landsberg/Lech, Germany.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

