Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex
- PMID: 18378696
- PMCID: PMC2423306
- DOI: 10.1128/MCB.01828-07
Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex
Abstract
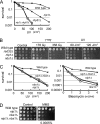
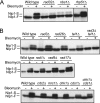
The Schizosaccharomyces pombe nip1(+)/ctp1(+) gene was previously identified as an slr (synthetically lethal with rad2) mutant. Epistasis analysis indicated that Nip1/Ctp1 functions in Rhp51-dependent recombinational repair, together with the Rad32 (spMre11)-Rad50-Nbs1 complex, which plays important roles in the early steps of DNA double-strand break repair. Nip1/Ctp1 was phosphorylated in asynchronous, exponentially growing cells and further phosphorylated in response to bleomycin treatment. Overproduction of Nip1/Ctp1 suppressed the DNA repair defect of an nbs1-s10 mutant, which carries a mutation in the FHA phosphopeptide-binding domain of Nbs1, but not of an nbs1 null mutant. Meiotic DNA double-strand breaks accumulated in the nip1/ctp1 mutant. The DNA repair phenotypes and epistasis relationships of nip1/ctp1 are very similar to those of the Saccharomyces cerevisiae sae2/com1 mutant, suggesting that Nip1/Ctp1 is a functional homologue of Sae2/Com1, although the sequence similarity between the proteins is limited to the C-terminal region containing the RHR motif. We found that the RxxL and CxxC motifs are conserved in Schizosaccharomyces species and in vertebrate CtIP, originally identified as a cofactor of the transcriptional corepressor CtBP. However, these two motifs are not found in other fungi, including Saccharomyces and Aspergillus species. We propose that Nip1/Ctp1 is a functional counterpart of Sae2/Com1 and CtIP.
Figures




References
-
- Alani, E., R. Padmore, and N. Kleckner. 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61419-436. - PubMed
-
- Arora, C., K. Kee, S. Maleki, and S. Keeney. 2004. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell 13549-559. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
