Progesterone induction of the 11beta-hydroxysteroid dehydrogenase type 2 promoter in breast cancer cells involves coordinated recruitment of STAT5A and progesterone receptor to a distal enhancer and polymerase tracking
- PMID: 18378698
- PMCID: PMC2423295
- DOI: 10.1128/MCB.01217-07
Progesterone induction of the 11beta-hydroxysteroid dehydrogenase type 2 promoter in breast cancer cells involves coordinated recruitment of STAT5A and progesterone receptor to a distal enhancer and polymerase tracking
Abstract
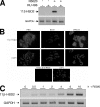
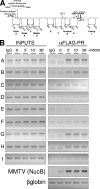
Steroid hormone receptors regulate gene expression, interacting with target DNA sequences but also activating cytoplasmic signaling pathways. Using the human 11beta-hydroxysteroid dehydrogenase type 2 (11beta-HSD2) gene as a model, we have investigated the contributions of both effects on a human progesterone-responsive promoter in breast cancer cells. Chromatin immunoprecipitation has identified two different mechanisms of hormone-induced progesterone receptor (PR) recruitment to the 11beta-HSD2 promoter: (i) direct PR binding to DNA at the proximal promoter, abrogated when PR contains a mutated DNA binding domain (DBD), and (ii) STAT5A (signal transducer and activator of transcription 5A)-mediated recruitment of PR to an upstream distal region, impaired by AG490, a JAK/STAT pathway inhibitor. The JAK/STAT inhibitor, as well as expression of dominant-negative STAT5A, impairs hormone induction of 11beta-HSD2. On the other hand, the DBD-mutated PR fully supports 11beta-HSD2 expression. These results, along with data from a deletion analysis, indicate that the distal region is crucial for hormone regulation of 11beta-HSD2. We show active RNA polymerase II tracking from the distal region upon PR and STAT5A binding, concomitant with synthesis of noncoding, hormone-dependent RNAs, suggesting that this region works as a hormone-dependent transcriptional enhancer. In conclusion, coordination of PR transcriptional effects and cytoplasmic signaling activation, in particular the JAK/STAT pathway, are critical in regulating progestin-induced endogenous 11beta-HSD2 gene expression in breast cancer cells. This is not unique to this promoter, as AG490 also alters the expression of other progesterone-regulated genes.
Figures








Similar articles
-
A Common Docking Domain in Progesterone Receptor-B links DUSP6 and CK2 signaling to proliferative transcriptional programs in breast cancer cells.Nucleic Acids Res. 2013 Oct;41(19):8926-42. doi: 10.1093/nar/gkt706. Epub 2013 Aug 5. Nucleic Acids Res. 2013. PMID: 23921636 Free PMC article.
-
Progesterone receptor directly inhibits β-casein gene transcription in mammary epithelial cells through promoting promoter and enhancer repressive chromatin modifications.Mol Endocrinol. 2011 Jun;25(6):955-68. doi: 10.1210/me.2011-0064. Epub 2011 Apr 28. Mol Endocrinol. 2011. PMID: 21527503 Free PMC article.
-
Progesterone receptor and Stat5 signaling cross talk through RANKL in mammary epithelial cells.Mol Endocrinol. 2013 Nov;27(11):1808-24. doi: 10.1210/me.2013-1077. Epub 2013 Sep 6. Mol Endocrinol. 2013. PMID: 24014651 Free PMC article.
-
Chromatin remodeling and control of cell proliferation by progestins via cross talk of progesterone receptor with the estrogen receptors and kinase signaling pathways.Ann N Y Acad Sci. 2006 Nov;1089:59-72. doi: 10.1196/annals.1386.025. Ann N Y Acad Sci. 2006. PMID: 17261755 Review.
-
More help than hindrance: nucleosomes aid transcriptional regulation.Nucleus. 2013 May-Jun;4(3):189-94. doi: 10.4161/nucl.25108. Epub 2013 May 28. Nucleus. 2013. PMID: 23756349 Free PMC article. Review.
Cited by
-
Transcriptional complexity and roles of Fra-1/AP-1 at the uPA/Plau locus in aggressive breast cancer.Nucleic Acids Res. 2014;42(17):11011-24. doi: 10.1093/nar/gku814. Epub 2014 Sep 8. Nucleic Acids Res. 2014. PMID: 25200076 Free PMC article.
-
A Common Docking Domain in Progesterone Receptor-B links DUSP6 and CK2 signaling to proliferative transcriptional programs in breast cancer cells.Nucleic Acids Res. 2013 Oct;41(19):8926-42. doi: 10.1093/nar/gkt706. Epub 2013 Aug 5. Nucleic Acids Res. 2013. PMID: 23921636 Free PMC article.
-
Research resource: progesterone receptor targetome underlying mammary gland branching morphogenesis.Mol Endocrinol. 2013 Oct;27(10):1743-61. doi: 10.1210/me.2013-1144. Epub 2013 Aug 26. Mol Endocrinol. 2013. PMID: 23979845 Free PMC article.
-
Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth.PLoS Genet. 2008 Oct;4(10):e1000227. doi: 10.1371/journal.pgen.1000227. Epub 2008 Oct 17. PLoS Genet. 2008. PMID: 18927631 Free PMC article.
-
Impact of chromatin structure on PR signaling: transition from local to global analysis.Mol Cell Endocrinol. 2012 Jun 24;357(1-2):30-6. doi: 10.1016/j.mce.2011.09.006. Epub 2011 Sep 21. Mol Cell Endocrinol. 2012. PMID: 21958695 Free PMC article. Review.
References
-
- Agarwal, A. K., and P. C. White. 1996. Analysis of the promoter of the NAD+ dependent 11 beta-hydroxysteroid dehydrogenase (HSD11K) gene in JEG-3 human choriocarcinoma cells. Mol. Cell. Endocrinol. 12193-99. - PubMed
-
- Albiston, A. L., V. R. Obeyesekere, R. E. Smith, and Z. S. Krozowski. 1994. Cloning and tissue distribution of the human 11 beta-hydroxysteroid dehydrogenase type 2 enzyme. Mol. Cell. Endocrinol. 105R11-R17. - PubMed
-
- Ariyoshi, K., T. Nosaka, K. Yamada, M. Onishi, Y. Oka, A. Miyajima, and T. Kitamura. 2000. Constitutive activation of STAT5 by a point mutation in the SH2 domain. J. Biol. Chem. 27524407-24413. - PubMed
-
- Ballaré, C., M. Uhrig, T. Bechtold, E. Sancho, M. Di Domenico, A. Migliaccio, F. Auricchio, and M. Beato. 2003. Two domains of the progesterone receptor interact with the estrogen receptor and are required for progesterone activation of the c-Src/Erk pathway in mammalian cells. Mol. Cell. Biol. 231994-2008. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
