In vivo study of the nucleosome assembly functions of ASF1 histone chaperones in human cells
- PMID: 18378699
- PMCID: PMC2423286
- DOI: 10.1128/MCB.00510-07
In vivo study of the nucleosome assembly functions of ASF1 histone chaperones in human cells
Abstract
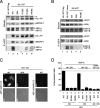
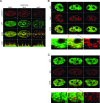
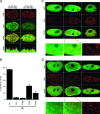
Histone chaperones have been implicated in nucleosome assembly and disassembly as well as histone modification. ASF1 is a highly conserved histone H3/H4 chaperone that synergizes in vitro with two other histone chaperones, chromatin assembly factor 1 (CAF-1) and histone repression A factor (HIRA), in DNA synthesis-coupled and DNA synthesis-independent nucleosome assembly. Here, we identify mutants of histones H3.1 and H3.3 that are unable to interact with human ASF1A and ASF1B isoforms but that are still competent to bind CAF-1 and HIRA, respectively. We show that these mutant histones are inefficiently deposited into chromatin in vivo. Furthermore, we found that both ASF1A and ASF1B participate in the DNA synthesis-independent deposition of H3.3 in HeLa cells, thus highlighting an unexpected role for ASF1B in this pathway. This pathway does not require interaction of ASF1 with HIRA. We provide the first direct determination that ASF1A and ASF1B play a role in the efficiency of nucleosome assembly in vivo in human cells.
Figures



References
-
- Adkins, M. W., and J. K. Tyler. 2004. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem. 27952069-52074. - PubMed
-
- Agez, M., J. Chen, R. Guerois, C. van Heijenoort, J. Y. Thuret, C. Mann, and F. Ochsenbein. 2007. Structure of the histone chaperone ASF1 bound to the histone H3 C-terminal helix and functional insights. Structure 15191-199. - PubMed
-
- Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 91191-1200. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
