"New-version-fast-multipole-method" accelerated electrostatic interactions in biomolecular systems
- PMID: 18379638
- PMCID: PMC2084081
- DOI: 10.1016/j.jcp.2007.05.026
"New-version-fast-multipole-method" accelerated electrostatic interactions in biomolecular systems
Abstract
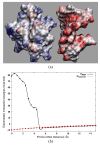
In this paper, we present an efficient and accurate numerical algorithm for calculating the electrostatic interactions in biomolecular systems. In our scheme, a boundary integral equation (BIE) approach is applied to discretize the linearized Poisson-Boltzmann (PB) equation. The resulting integral formulas are well conditioned for single molecule cases as well as for systems with more than one macromolecule, and are solved efficiently using Krylov subspace based iterative methods such as generalized minimal residual (GMRES) or bi-conjugate gradients stabilized (BiCGStab) methods. In each iteration, the convolution type matrix-vector multiplications are accelerated by a new version of the fast multipole method (FMM). The implemented algorithm is asymptotically optimal O(N) both in CPU time and memory usage with optimized prefactors. Our approach enhances the present computational ability to treat electrostatics of large scale systems in protein-protein interactions and nano particle assembly processes. Applications including calculating the electrostatics of the nicotinic acetylcholine receptor (nAChR) and interactions between protein Sso7d and DNA are presented.
Figures





Similar articles
-
AFMPB: An Adaptive Fast Multipole Poisson-Boltzmann Solver for Calculating Electrostatics in Biomolecular Systems.Comput Phys Commun. 2010 Jun 1;181(6):1150-1160. doi: 10.1016/j.cpc.2010.02.015. Comput Phys Commun. 2010. PMID: 20532187 Free PMC article.
-
Order N algorithm for computation of electrostatic interactions in biomolecular systems.Proc Natl Acad Sci U S A. 2006 Dec 19;103(51):19314-9. doi: 10.1073/pnas.0605166103. Epub 2006 Dec 5. Proc Natl Acad Sci U S A. 2006. PMID: 17148613 Free PMC article.
-
An Adaptive Fast Multipole Boundary Element Method for Poisson-Boltzmann Electrostatics.J Chem Theory Comput. 2009 Jun 9;5(6):1692-1699. doi: 10.1021/ct900083k. Epub 2009 May 21. J Chem Theory Comput. 2009. PMID: 19517026 Free PMC article.
-
Profiling General-Purpose Fast Multipole Method (FMM) Using Human Head Topology.2020 Aug 6. In: Makarov SN, Noetscher GM, Nummenmaa A, editors. Brain and Human Body Modeling 2020: Computational Human Models Presented at EMBC 2019 and the BRAIN Initiative® 2019 Meeting [Internet]. Cham (CH): Springer; 2021. 2020 Aug 6. In: Makarov SN, Noetscher GM, Nummenmaa A, editors. Brain and Human Body Modeling 2020: Computational Human Models Presented at EMBC 2019 and the BRAIN Initiative® 2019 Meeting [Internet]. Cham (CH): Springer; 2021. PMID: 32966013 Free Books & Documents. Review.
-
Polarizable atomic multipole solutes in a Poisson-Boltzmann continuum.J Chem Phys. 2007 Mar 28;126(12):124114. doi: 10.1063/1.2714528. J Chem Phys. 2007. PMID: 17411115 Free PMC article. Review.
Cited by
-
A Sixth-order Image Approximation to the Ionic Solvent Induced Reaction Field.J Sci Comput. 2009 Dec 1;41(3):411-435. doi: 10.1007/s10915-009-9307-z. J Sci Comput. 2009. PMID: 21152236 Free PMC article.
-
AFMPB: An Adaptive Fast Multipole Poisson-Boltzmann Solver for Calculating Electrostatics in Biomolecular Systems.Comput Phys Commun. 2010 Jun 1;181(6):1150-1160. doi: 10.1016/j.cpc.2010.02.015. Comput Phys Commun. 2010. PMID: 20532187 Free PMC article.
-
RETRACTED: Capacitive Behavior of Aqueous Electrical Double Layer Based on Dipole Dimer Water Model.Nanomaterials (Basel). 2022 Dec 20;13(1):16. doi: 10.3390/nano13010016. Nanomaterials (Basel). 2022. Retraction in: Nanomaterials (Basel). 2024 Dec 24;15(1):1. doi: 10.3390/nano15010001. PMID: 36615925 Free PMC article. Retracted.
-
A Fast and Robust Poisson-Boltzmann Solver Based on Adaptive Cartesian Grids.J Chem Theory Comput. 2011 May 10;7(5):1524-1540. doi: 10.1021/ct1006983. J Chem Theory Comput. 2011. PMID: 21984876 Free PMC article.
-
The de Rham-Hodge Analysis and Modeling of Biomolecules.Bull Math Biol. 2020 Aug 8;82(8):108. doi: 10.1007/s11538-020-00783-2. Bull Math Biol. 2020. PMID: 32770408 Free PMC article.
References
-
- Abramowitz M, Stegun IA. Handbook of Mathematical Functions. Dover Publications; New York: 1965.
-
- Altman M, Bardhan J, White J, Tidor B. An accurate surface formulation for biomolecule electrostatics in non-ionic solutions. Conf Proc IEEE Eng Med Biol Soc. 2005;7(NIL):7591–5. - PubMed
-
- Altman MD, Bardhan JP, Tidor B, White JK. FFTSVD: a fast multiscale boundary-element method solver suitable for Bio-MEMS and biomolecule simulation. IEEE Trans Comput-Aided Des Integr Circuits Syst. 2006;25(2):274–284.
-
- Appel AW. An efficient program for many-body simulations. SIAM J Sci Stat Comput. 1985;6:85–103.
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
