Appearance and propagation of polyglutamine-based amyloids in yeast: tyrosine residues enable polymer fragmentation
- PMID: 18381282
- PMCID: PMC2397454
- DOI: 10.1074/jbc.M802071200
Appearance and propagation of polyglutamine-based amyloids in yeast: tyrosine residues enable polymer fragmentation
Abstract
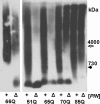
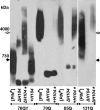
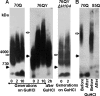
In yeast, fragmentation of amyloid polymers by the Hsp104 chaperone allows them to propagate as prions. The prion-forming domain of the yeast Sup35 protein is rich in glutamine, asparagine, tyrosine, and glycine residues, which may define its prion properties. Long polyglutamine stretches can also drive amyloid polymerization in yeast, but these polymers are unable to propagate because of poor fragmentation and exist through constant seeding with the Rnq1 prion polymers. We proposed that fragmentation of polyglutamine amyloids may be improved by incorporation of hydrophobic amino acid residues into polyglutamine stretches. To investigate this, we constructed sets of polyglutamine with or without tyrosine stretches fused to the non-prion domains of Sup35. Polymerization of these chimeras started rapidly, and its efficiency increased with stretch size. Polymerization of proteins with polyglutamine stretches shorter than 70 residues required Rnq1 prion seeds. Proteins with longer stretches polymerized independently of Rnq1 and thus could propagate. The presence of tyrosines within polyglutamine stretches dramatically enhanced polymer fragmentation and allowed polymer propagation in the absence of Rnq1 and, in some cases, of Hsp104.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

