DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes
- PMID: 18381894
- PMCID: PMC2279202
- DOI: 10.1101/gad.1640708
DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes
Abstract
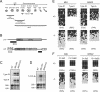
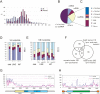
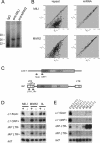
Silencing of transposable elements occurs during fetal gametogenesis in males via de novo DNA methylation of their regulatory regions. The loss of MILI (miwi-like) and MIWI2 (mouse piwi 2), two mouse homologs of Drosophila Piwi, activates retrotransposon gene expression by impairing DNA methylation in the regulatory regions of the retrotransposons. However, as it is unclear whether the defective DNA methylation in the mutants is due to the impairment of de novo DNA methylation, we analyze DNA methylation and Piwi-interacting small RNA (piRNA) expression in wild-type, MILI-null, and MIWI2-null male fetal germ cells. We reveal that defective DNA methylation of the regulatory regions of the Line-1 (long interspersed nuclear elements) and IAP (intracisternal A particle) retrotransposons in the MILI-null and MIWI2-null male germ cells takes place at the level of de novo methylation. Comprehensive analysis shows that the piRNAs of fetal germ cells are distinct from those previously identified in neonatal and adult germ cells. The expression of piRNAs is reduced under MILI- and MIWI2-null conditions in fetal germ cells, although the extent of the reduction differs significantly between the two mutants. Our data strongly suggest that MILI and MIWI2 play essential roles in establishing de novo DNA methylation of retrotransposons in fetal male germ cells.
Figures

Comment in
-
Small RNA guides for de novo DNA methylation in mammalian germ cells.Genes Dev. 2008 Apr 15;22(8):970-5. doi: 10.1101/gad.1669408. Genes Dev. 2008. PMID: 18413711 Free PMC article.
References
-
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
-
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. - PubMed
-
- Bourc’his D., Bestor T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. - PubMed
-
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
