Protein networks in disease
- PMID: 18381899
- PMCID: PMC3863981
- DOI: 10.1101/gr.071852.107
Protein networks in disease
Abstract
During a decade of proof-of-principle analysis in model organisms, protein networks have been used to further the study of molecular evolution, to gain insight into the robustness of cells to perturbation, and for assignment of new protein functions. Following these analyses, and with the recent rise of protein interaction measurements in mammals, protein networks are increasingly serving as tools to unravel the molecular basis of disease. We review promising applications of protein networks to disease in four major areas: identifying new disease genes; the study of their network properties; identifying disease-related subnetworks; and network-based disease classification. Applications in infectious disease, personalized medicine, and pharmacology are also forthcoming as the available protein network information improves in quality and coverage.
Figures

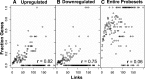
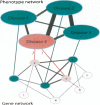
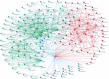
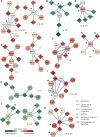
References
-
- Alon U. Introduction to systems biology: Design principles of biological circuits. Chapman and Hall; London, UK: 2006.
-
- Bader J.S., Chaudhuri A., Rothberg J.M., Chant J. Gaining confidence in high-throughput protein interaction networks. Nat. Biotechnol. 2004;22:78–85. - PubMed
-
- Barabasi A.L., Oltvai Z.N. Network biology: Understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
