Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C)
- PMID: 18382652
- PMCID: PMC2268738
- DOI: 10.1371/journal.pone.0001847
Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C)
Abstract
Background: Human rhinoviruses (HRVs) are the most frequently detected pathogens in acute respiratory tract infections (ARTIs) and yet little is known about the prevalence, recurrence, structure and clinical impact of individual members. During 2007, the complete coding sequences of six previously unknown and highly divergent HRV strains were reported. To catalogue the molecular and clinical features distinguishing the divergent HRV strains, we undertook, for the first time, in silico analyses of all available polyprotein sequences and performed retrospective reviews of the medical records of cases in which variants of the prototype strain, HRV-QPM, had been detected.
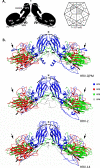
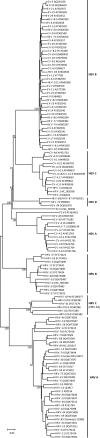
Methodology/principle findings: Genomic analyses revealed that the six divergent strains, residing within a clade we previously called HRV A2, had the shortest polyprotein of all picornaviruses investigated. Structure-based amino acid alignments identified conserved motifs shared among members of the genus Rhinovirus as well as substantive deletions and insertions unique to the divergent strains. Deletions mostly affected regions encoding proteins traditionally involved in antigenicity and serving as HRV and HEV receptor footprints. Because the HRV A2 strains cannot yet be cultured, we created homology models of predicted HRV-QPM structural proteins. In silico comparisons confirmed that HRV-QPM was most closely related to the major group HRVs. HRV-QPM was most frequently detected in infants with expiratory wheezing or persistent cough who had been admitted to hospital and required supplemental oxygen. It was the only virus detected in 65% of positive individuals. These observations contributed to an objective clinical impact ranging from mild to severe.
Conclusions: The divergent strains did not meet classification requirements for any existing species of the genus Rhinovirus or Enterovirus. HRV A2 strains should be partitioned into at least one new species, putatively called Human rhinovirus C, populated by members detected with high frequency, from individuals with respiratory symptoms requiring hospital admission.
Conflict of interest statement
Figures




Similar articles
-
Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis.J Clin Virol. 2007 Jun;39(2):67-75. doi: 10.1016/j.jcv.2007.03.012. Epub 2007 May 7. J Clin Virol. 2007. PMID: 17482871 Free PMC article.
-
Phylogenetic analysis of human rhinovirus capsid protein VP1 and 2A protease coding sequences confirms shared genus-like relationships with human enteroviruses.J Gen Virol. 2005 Mar;86(Pt 3):697-706. doi: 10.1099/vir.0.80445-0. J Gen Virol. 2005. PMID: 15722530
-
Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children.J Clin Microbiol. 2007 Nov;45(11):3655-64. doi: 10.1128/JCM.01254-07. Epub 2007 Sep 5. J Clin Microbiol. 2007. PMID: 17804649 Free PMC article.
-
Newly identified human rhinoviruses: molecular methods heat up the cold viruses.Rev Med Virol. 2010 May;20(3):156-76. doi: 10.1002/rmv.644. Rev Med Virol. 2010. PMID: 20127751 Free PMC article. Review.
-
Proposals for the classification of human rhinovirus species C into genotypically assigned types.J Gen Virol. 2010 Oct;91(Pt 10):2409-19. doi: 10.1099/vir.0.023994-0. Epub 2010 Jul 7. J Gen Virol. 2010. PMID: 20610666 Review.
Cited by
-
Year-Long Rhinovirus Infection is Influenced by Atmospheric Conditions, Outdoor Air Virus Presence, and Immune System-Related Genetic Polymorphisms.Food Environ Virol. 2019 Dec;11(4):340-349. doi: 10.1007/s12560-019-09397-x. Epub 2019 Jul 26. Food Environ Virol. 2019. PMID: 31350695
-
All known human rhinovirus species are present in sputum specimens of military recruits during respiratory infection.Viruses. 2009 Dec;1(3):1178-89. doi: 10.3390/v1031178. Epub 2009 Dec 4. Viruses. 2009. PMID: 21994588 Free PMC article.
-
Complete coding sequence characterization and comparative analysis of the putative novel human rhinovirus (HRV) species C and B.Virol J. 2011 Jan 7;8:5. doi: 10.1186/1743-422X-8-5. Virol J. 2011. PMID: 21214911 Free PMC article.
-
Pneumonia and pericarditis in a child with HRV-C infection: a case report.J Clin Virol. 2009 Jun;45(2):157-60. doi: 10.1016/j.jcv.2009.03.014. Epub 2009 May 7. J Clin Virol. 2009. PMID: 19427260 Free PMC article.
-
The role of rhinovirus infections in the development of early childhood asthma.Curr Opin Allergy Clin Immunol. 2010 Apr;10(2):133-8. doi: 10.1097/ACI.0b013e3283352f7c. Curr Opin Allergy Clin Immunol. 2010. PMID: 19996738 Free PMC article. Review.
References
-
- Kiang D, Yagi S, Kantardjieff KA, Kim EJ, Louie JK, et al. Molecular characterization of a variant rhinovirus from an outbreak associated with uncommonly high mortality. J Clin Virol. 2007;38:227–237. - PubMed
-
- Papadopoulos NG, Bates PJ, Bardin PG, Papi A, Leir SH, et al. Rhinoviruses infect the lower airways. J Infect Dis. 2000;181:1875–1884. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

