Biofilm induced tolerance towards antimicrobial peptides
- PMID: 18382672
- PMCID: PMC2270907
- DOI: 10.1371/journal.pone.0001891
Biofilm induced tolerance towards antimicrobial peptides
Abstract
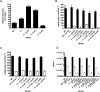
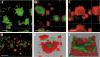
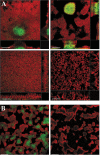
Increased tolerance to antimicrobial agents is thought to be an important feature of microbes growing in biofilms. We address the question of how biofilm organization affects antibiotic susceptibility. We established Escherichia coli biofilms with differential structural organization due to the presence of IncF plasmids expressing altered forms of the transfer pili in two different biofilm model systems. The mature biofilms were subsequently treated with two antibiotics with different molecular targets, the peptide antibiotic colistin and the fluoroquinolone ciprofloxacin. The dynamics of microbial killing were monitored by viable count determination, and confocal laser microscopy. Strains forming structurally organized biofilms show an increased bacterial survival when challenged with colistin, compared to strains forming unstructured biofilms. The increased survival is due to genetically regulated tolerant subpopulation formation and not caused by a general biofilm property. No significant difference in survival was detected when the strains were challenged with ciprofloxacin. Our data show that biofilm formation confers increased colistin tolerance to cells within the biofilm structure, but the protection is conditional being dependent on the structural organization of the biofilm, and the induction of specific tolerance mechanisms.
Conflict of interest statement
Figures
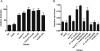
Similar articles
-
Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes.Mol Microbiol. 2008 Apr;68(1):223-40. doi: 10.1111/j.1365-2958.2008.06152.x. Epub 2008 Feb 28. Mol Microbiol. 2008. PMID: 18312276
-
Effects of colistin on biofilm matrices of Escherichia coli and Staphylococcus aureus.Int J Antimicrob Agents. 2017 Apr;49(4):472-479. doi: 10.1016/j.ijantimicag.2017.01.005. Epub 2017 Mar 4. Int J Antimicrob Agents. 2017. PMID: 28267594
-
Effects of the twin-arginine translocase on the structure and antimicrobial susceptibility of Escherichia coli biofilms.Can J Microbiol. 2005 Aug;51(8):671-83. doi: 10.1139/w05-048. Can J Microbiol. 2005. PMID: 16234865
-
[Current progress in antimicrobial peptides against bacterial biofilms].Sheng Wu Gong Cheng Xue Bao. 2020 Jul 25;36(7):1277-1282. doi: 10.13345/j.cjb.190511. Sheng Wu Gong Cheng Xue Bao. 2020. PMID: 32748585 Review. Chinese.
-
Use of antimicrobial peptides against microbial biofilms: advantages and limits.Curr Med Chem. 2011;18(2):256-79. doi: 10.2174/092986711794088399. Curr Med Chem. 2011. PMID: 21110801 Review.
Cited by
-
Estimation of a biofilm-specific reaction rate: kinetics of bacterial urea hydrolysis in a biofilm.NPJ Biofilms Microbiomes. 2015 Sep 16;1:15014. doi: 10.1038/npjbiofilms.2015.14. eCollection 2015. NPJ Biofilms Microbiomes. 2015. PMID: 28721232 Free PMC article.
-
Real-time solvent tolerance analysis of pseudomonas sp. strain VLB120{Delta}C catalytic biofilms.Appl Environ Microbiol. 2011 Mar;77(5):1563-71. doi: 10.1128/AEM.02498-10. Epub 2010 Dec 30. Appl Environ Microbiol. 2011. PMID: 21193676 Free PMC article.
-
Experimental and Theoretical Investigation of Multispecies Oral Biofilm Resistance to Chlorhexidine Treatment.Sci Rep. 2016 Jun 21;6:27537. doi: 10.1038/srep27537. Sci Rep. 2016. PMID: 27325010 Free PMC article.
-
The peptidoglycan and biofilm matrix of Staphylococcus epidermidis undergo structural changes when exposed to human platelets.PLoS One. 2019 Jan 25;14(1):e0211132. doi: 10.1371/journal.pone.0211132. eCollection 2019. PLoS One. 2019. PMID: 30682094 Free PMC article.
-
Saccharomyces cerevisiae biofilm tolerance towards systemic antifungals depends on growth phase.BMC Microbiol. 2014 Dec 4;14:305. doi: 10.1186/s12866-014-0305-4. BMC Microbiol. 2014. PMID: 25472667 Free PMC article.
References
-
- Ghigo JM. Natural conjugative plasmids induce bacterial biofilm development. Nature. 2001;412:442–445. - PubMed
-
- Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol. 2003;14:255–261. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

