Quantitative multicolor compositional imaging resolves molecular domains in cell-matrix adhesions
- PMID: 18382676
- PMCID: PMC2270910
- DOI: 10.1371/journal.pone.0001901
Quantitative multicolor compositional imaging resolves molecular domains in cell-matrix adhesions
Abstract
Background: Cellular processes occur within dynamic and multi-molecular compartments whose characterization requires analysis at high spatio-temporal resolution. Notable examples for such complexes are cell-matrix adhesion sites, consisting of numerous cytoskeletal and signaling proteins. These adhesions are highly variable in their morphology, dynamics, and apparent function, yet their molecular diversity is poorly defined.
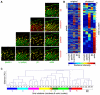
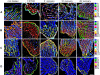
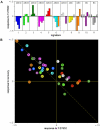
Methodology/principal findings: We present here a compositional imaging approach for the analysis and display of multi-component compositions. This methodology is based on microscopy-acquired multicolor data, multi-dimensional clustering of pixels according to their composition similarity and display of the cellular distribution of these composition clusters. We apply this approach for resolving the molecular complexes associated with focal-adhesions, and the time-dependent effects of Rho-kinase inhibition. We show here compositional variations between adhesion sites, as well as ordered variations along the axis of individual focal-adhesions. The multicolor clustering approach also reveals distinct sensitivities of different focal-adhesion-associated complexes to Rho-kinase inhibition.
Conclusions/significance: Multicolor compositional imaging resolves "molecular signatures" characteristic to focal-adhesions and related structures, as well as sub-domains within these adhesion sites. This analysis enhances the spatial information with additional "contents-resolved" dimensions. We propose that compositional imaging can serve as a powerful tool for studying complex multi-molecular assemblies in cells and for mapping their distribution at sub-micron resolution.
Conflict of interest statement
Figures


References
-
- Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, et al. Single Cell Profiling of Potentiated Phospho-Protein Networks in Cancer Cells. Cell. 2004;118:217–228. - PubMed
-
- De Rosa SC, Herzenberg LA, Roederer M. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat Med. 2001;7:245–248. - PubMed
-
- Mallick P, Schirle M, Chen SS, Flory MR, Lee H, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25:125–131. - PubMed
-
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. - PubMed
-
- Rhodes DR, Chinnaiyan AM. Integrative analysis of the cancer transcriptome. Nat Genet 37 Suppl. 2005:S31–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

