Alignment of non-covalent interactions at protein-protein interfaces
- PMID: 18382693
- PMCID: PMC2274958
- DOI: 10.1371/journal.pone.0001926
Alignment of non-covalent interactions at protein-protein interfaces
Abstract
Background: The study and comparison of protein-protein interfaces is essential for the understanding of the mechanisms of interaction between proteins. While there are many methods for comparing protein structures and protein binding sites, so far no methods have been reported for comparing the geometry of non-covalent interactions occurring at protein-protein interfaces.
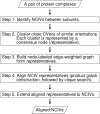
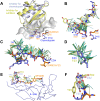
Methodology/principal findings: Here we present a method for aligning non-covalent interactions between different protein-protein interfaces. The method aligns the vector representations of van der Waals interactions and hydrogen bonds based on their geometry. The method has been applied to a dataset which comprises a variety of protein-protein interfaces. The alignments are consistent to a large extent with the results obtained using two other complementary approaches. In addition, we apply the method to three examples of protein mimicry. The method successfully aligns respective interfaces and allows for recognizing conserved interface regions.
Conclusions/significance: The Galinter method has been validated in the comparison of interfaces in which homologous subunits are involved, including cases of mimicry. The method is also applicable to comparing interfaces involving non-peptidic compounds. Galinter assists users in identifying local interface regions with similar patterns of non-covalent interactions. This is particularly relevant to the investigation of the molecular basis of interaction mimicry.
Conflict of interest statement
Figures

References
-
- Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285(5):2177–98. - PubMed
-
- Sheinerman FB, Norel R, Honig B. Electrostatic aspects of protein-protein interactions. Curr Opin Struct Biol. 2000;10(2):153–159. - PubMed
-
- Rodier F, Bahadur RP, Chakrabarti P, Janin J. Hydration of protein-protein interfaces. Proteins. 2005;60:36–45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

