Refinement of noncalorimetric determination of the change in heat capacity, DeltaC(p), of protein unfolding and validation across a wide temperature range
- PMID: 18384147
- PMCID: PMC2630543
- DOI: 10.1002/prot.22016
Refinement of noncalorimetric determination of the change in heat capacity, DeltaC(p), of protein unfolding and validation across a wide temperature range
Abstract
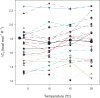
The change in heat capacity, DeltaC(p), on protein unfolding has been usually determined by calorimetry. A noncalorimetric method which employs the Gibbs-Helmholtz relationship to determine DeltaC(p) has seen some use. Generally, in this method the free energy change on unfolding of the protein is determined at a variety of temperatures and the temperature at which DeltaG is zero, T(m), and change in enthalpy at T(m) are determined by thermal denaturation and DeltaC(p) is then calculated using the Gibbs-Helmholtz equation. We show here that an abbreviated method with stability determinations at just two temperatures gives values of DeltaC(p) consistent with values from free energy change on unfolding determination at a much wider range of temperatures. Further, even the free energy change on unfolding from a single solvent denaturation at the proper temperature, when coupled with the melting temperature, T(m), and the van't Hoff enthalpy, DeltaH(vH), from a thermal denaturation, gives a reasonable estimate of DeltaC(p), albeit with greater uncertainty than solvent denaturations at two temperatures. We also find that nonlinear regression of the Gibbs-Helmholtz equation as a function of stability and temperature while simultaneously fitting DeltaC(p), T(m), and DeltaH(vH) gives values for the last two parameters that are in excellent agreement with experimental values.
(c) 2008 Wiley-Liss, Inc.
Figures





References
-
- Pace CN, Laurents DV. A new method for determining the heat capacity change for protein folding. Biochemistry. 1989;28:2520–2525. - PubMed
-
- Schwehm JM, Stites WE. Application of automated methods for determination of protein conformational stability. Methods Enzymol. 1998;295:150–170. - PubMed
-
- Stites WE, Byrne MP, Aviv J, Kaplan M, Curtis PM. Instrumentation for automated determination of protein stability. Anal Biochem. 1995;227:112–122. - PubMed
-
- Holder JB, Bennett AF, Chen J, Spencer DS, Byrne MP, Stites WE. Energetics of side chain packing in staphylococcal nuclease assessed by exchange of valines, isoleucines, and leucines. Biochemistry. 2001;40:13998–14003. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

