Novel method for probing the specificity binding profile of ligands: applications to HIV protease
- PMID: 18384529
- PMCID: PMC3480184
- DOI: 10.1111/j.1747-0285.2008.00659.x
Novel method for probing the specificity binding profile of ligands: applications to HIV protease
Abstract
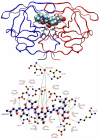
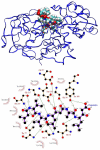
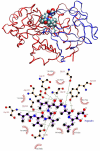
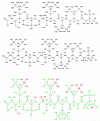
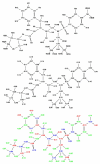
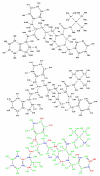
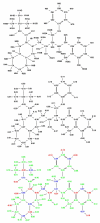
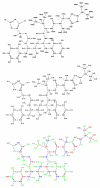
A detailed understanding of factors influencing the binding specificity of a ligand to a set of desirable targets and undesirable decoys is a key step in the design of potent and selective therapeutics. We have developed a general method for optimizing binding specificity in ligand-receptor complexes based on the theory of electrostatic charge optimization. This methodology can be used to tune the binding of a ligand to a panel of potential targets and decoys, along the continuum from narrow binding to only one partner to broad binding to the entire panel. Using HIV-1 protease as a model system, we probe specificity in three distinct ways. First, we probe interactions that could make the promiscuous protease inhibitor pepstatin more selective toward HIV-1 protease. Next, we study clinically approved HIV-1 protease inhibitors and probe ways to broaden the binding profiles toward both wild-type HIV-1 protease and drug-resistant mutants. Finally, we study a conformational ensemble of wild-type HIV-1 protease to 'design in' broad specificity to known drugs before resistance mutations arise. The results from this conformational ensemble were similar to those from the drug-resistant ensemble, suggesting the use of a conformational wild-type ensemble as a tool to develop escape-mutant-resistant inhibitors.
Figures
References
-
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(6761 Suppl):C47–C52. - PubMed
-
- Yeh P, Tschumi A, Kishony R. Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 2006;38:489–494. - PubMed
-
- Sebolt-Leopold J, English J. Mechanisms of drug inhibition of signalling molecules. Nature. 2006;441:457–462. - PubMed
-
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

