The kinetic and equilibrium molten globule intermediates of apoleghemoglobin differ in structure
- PMID: 18384808
- PMCID: PMC2409369
- DOI: 10.1016/j.jmb.2008.03.025
The kinetic and equilibrium molten globule intermediates of apoleghemoglobin differ in structure
Abstract
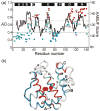
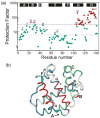
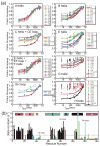
An important question in protein folding is whether molten globule states formed under equilibrium conditions are good structural models for kinetic folding intermediates. The structures of the kinetic and equilibrium intermediates in the folding of the plant globin apoleghemoglobin have been compared at high resolution by quench-flow pH-pulse labeling and interrupted hydrogen/deuterium exchange analyzed in dimethyl sulfoxide. Unlike its well studied homolog apomyoglobin, where the equilibrium and kinetic intermediates are quite similar, there are striking structural differences between the intermediates formed by apoleghemoglobin. In the kinetic intermediate, formed during the burst phase of the quench-flow experiment, protected amides and helical structure are found mainly in the regions corresponding to the G and H helices of the folded protein, and in parts of the E helix and CE loop regions, whereas in the equilibrium intermediate, amide protection and helical structure are seen in parts of the A and B helix regions, as well as in the G and H regions, and the E helix remains largely unfolded. These results suggest that the structure of the molten globule intermediate of apoleghemoglobin is more plastic than that of apomyoglobin, so that it is readily transformed depending on the solution conditions, particularly pH. Thus, in the case of apoleghemoglobin at least, the equilibrium molten globule formed under destabilizing conditions at acid pH is not a good model for the compact intermediate formed during kinetic refolding experiments. Our high-precision kinetic analysis also reveals an additional slow phase during the folding of apoleghemoglobin, which is not observed for apomyoglobin. Hydrogen exchange pulse-labeling experiments show that the slow-folding phase is associated with residues in the CE loop, which probably forms non-native structure in the intermediate that must be resolved before folding can proceed to completion.
Figures


Similar articles
-
Folding of apomyoglobin: Analysis of transient intermediate structure during refolding using quick hydrogen deuterium exchange and NMR.Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(1):10-27. doi: 10.2183/pjab.93.002. Proc Jpn Acad Ser B Phys Biol Sci. 2017. PMID: 28077807 Free PMC article. Review.
-
Conformational and dynamic characterization of the molten globule state of an apomyoglobin mutant with an altered folding pathway.Biochemistry. 2001 Dec 4;40(48):14459-67. doi: 10.1021/bi011500n. Biochemistry. 2001. PMID: 11724558
-
Identification of native and non-native structure in kinetic folding intermediates of apomyoglobin.J Mol Biol. 2006 Jan 6;355(1):139-56. doi: 10.1016/j.jmb.2005.10.047. Epub 2005 Nov 8. J Mol Biol. 2006. PMID: 16300787
-
Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin.Science. 1993 Nov 5;262(5135):892-6. doi: 10.1126/science.8235610. Science. 1993. PMID: 8235610
-
Non-Native Structures of Apomyoglobin and Apoleghemoglobin in Folding Intermediates Related to the Protein Misfolding.Molecules. 2023 May 8;28(9):3970. doi: 10.3390/molecules28093970. Molecules. 2023. PMID: 37175379 Free PMC article. Review.
Cited by
-
The use of spin desalting columns in DMSO-quenched H/D-exchange NMR experiments.Protein Sci. 2013 Apr;22(4):486-91. doi: 10.1002/pro.2221. Epub 2013 Feb 11. Protein Sci. 2013. PMID: 23339068 Free PMC article.
-
NMR-based characterization of a refolding intermediate of beta2-microglobulin labeled using a wheat germ cell-free system.Protein Sci. 2009 Aug;18(8):1592-601. doi: 10.1002/pro.179. Protein Sci. 2009. PMID: 19606503 Free PMC article.
-
Folding of apomyoglobin: Analysis of transient intermediate structure during refolding using quick hydrogen deuterium exchange and NMR.Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(1):10-27. doi: 10.2183/pjab.93.002. Proc Jpn Acad Ser B Phys Biol Sci. 2017. PMID: 28077807 Free PMC article. Review.
-
Interrupted hydrogen/deuterium exchange reveals the stable core of the remarkably helical molten globule of alpha-beta parallel protein flavodoxin.J Biol Chem. 2010 Feb 5;285(6):4165-4172. doi: 10.1074/jbc.M109.087932. Epub 2009 Dec 3. J Biol Chem. 2010. PMID: 19959481 Free PMC article.
-
Consequences of stabilizing the natively disordered f helix for the folding pathway of apomyoglobin.J Mol Biol. 2011 Aug 5;411(1):248-63. doi: 10.1016/j.jmb.2011.05.028. Epub 2011 May 27. J Mol Biol. 2011. PMID: 21640124 Free PMC article.
References
-
- Bashford D, Chothia C, Lesk AM. Determinants of a protein fold. Unique features of the globin amino acid sequence. J Mol Biol. 1987;196:199–216. - PubMed
-
- Appleby CA. Leghemoglobin and Rhizobium respiration. Ann Rev Plant Physiol. 1984;35:443–478.
-
- Appleby CA. The origin and functions of hemoglobin in plants. Sci Progr. 1992;76:365–398.
-
- Jennings PA, Wright PE. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science. 1993;262:892–896. - PubMed
-
- Nishimura C, Prytulla S, Dyson HJ, Wright PE. Conservation of folding pathways in evolutionarily distant globin sequences. Nat Struct Biol. 2000;7:679–686. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

