Minimal components of the RNA polymerase II transcription apparatus determine the consensus TATA box
- PMID: 18385157
- PMCID: PMC2396422
- DOI: 10.1093/nar/gkn130
Minimal components of the RNA polymerase II transcription apparatus determine the consensus TATA box
Abstract
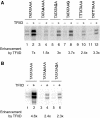
In Saccharomyces cerevisiae, multiple approaches have arrived at a consensus TATA box sequence of TATA(T/A)A(A/T)(A/G). TATA-binding protein (TBP) affinity alone does not determine TATA box function. To discover how a minimal set of factors required for basal and activated transcription contributed to the sequence requirements for a functional TATA box, we performed transcription reactions using highly purified proteins and CYC1 promoter TATA box mutants. The TATA box consensus sequence is a good predictor of promoter activity. However, several nonconsensus sequences are almost fully functional, indicating that mechanistic requirements are not the only selective pressure on the TATA box. We also found that the effect of a mutation at a certain position is often dependent on other bases within a particular TATA box. Although activators and coactivators strongly influence TBP recruitment and stability at promoters, neither Mediator, the activator Gal4-V16, nor TFIID specifically compensate for the low transcription levels of the weak TATA boxes. The addition of Mediator to purified transcription reactions did, however, increase the functional selectivity for certain consensus TATA sequences. Transcription in whole-cell extracts or in vivo with these TATA box mutants indicated that factors, other than those in our purified system, may help initiate transcription from weak TATA boxes.
Figures

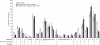




References
-
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. - PubMed
-
- Müller F, Demény MA, Tora L. New problems in RNA polymerase II transcription initiation: Matching the diversity of core promoters with a variety of promoter recognition factors. J. Biol. Chem. 2007;282:14685–14689. - PubMed
-
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

