Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes
- PMID: 18385237
- PMCID: PMC2493305
- DOI: 10.1128/JVI.00104-08
Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes
Figures


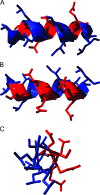
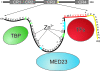
References
-
- Alevizopoulos, K., B. Sanchez, and B. Amati. 2000. Conserved region 2 of adenovirus E1A has a function distinct from pRb binding required to prevent cell cycle arrest by p16INK4a or p27Kip1. Oncogene 192067-2074. - PubMed
-
- Ansieau, S., and A. Leutz. 2002. The conserved Mynd domain of BS69 binds cellular and oncoviral proteins through a common PXLXP motif. J. Biol. Chem. 2774906-4910. - PubMed
-
- Arany, Z., D. Newsome, E. Oldread, D. M. Livingston, and R. Eckner. 1995. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 37481-84. - PubMed
-
- Avvakumov, N., A. E. Kajon, R. C. Hoeben, and J. S. Mymryk. 2004. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology 329477-492. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

