Comprehensive immunological evaluation reveals surprisingly few differences between elite controller and progressor Mamu-B*17-positive simian immunodeficiency virus-infected rhesus macaques
- PMID: 18385251
- PMCID: PMC2395202
- DOI: 10.1128/JVI.00292-08
Comprehensive immunological evaluation reveals surprisingly few differences between elite controller and progressor Mamu-B*17-positive simian immunodeficiency virus-infected rhesus macaques
Abstract
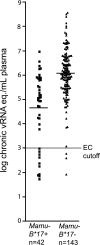
The association between particular major histocompatibility complex class I (MHC-I) alleles and control of human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) replication implies that certain CD8(+) T-lymphocyte (CD8-TL) responses are better able than others to control viral replication in vivo. However, possession of favorable alleles does not guarantee improved prognosis or viral control. In rhesus macaques, the MHC-I allele Mamu-B*17 is correlated with reduced viremia and is overrepresented in macaques that control SIVmac239, termed elite controllers (ECs). However, there is so far no mechanistic explanation for this phenomenon. Here we show that the chronic-phase Mamu-B*17-restricted repertoire is focused primarily against just five epitopes-VifHW8, EnvFW9, NefIW9, NefMW9, and env(ARF)cRW9-in both ECs and progressors. Interestingly, Mamu-B*17-restricted CD8-TL do not target epitopes in Gag. CD8-TL escape variation occurred in all targeted Mamu-B*17-restricted epitopes. However, recognition of escape variant peptides was commonly observed in both ECs and progressors. Wild-type sequences in the VifHW8 epitope tended to be conserved in ECs, but there was no evidence that this enhances viral control. In fact, no consistent differences were detected between ECs and progressors in any measured parameter. Our data suggest that the narrowly focused Mamu-B*17-restricted repertoire suppresses virus replication and drives viral evolution. It is, however, insufficient in the majority of individuals that express the "protective" Mamu-B*17 molecule. Most importantly, our data indicate that the important differences between Mamu-B*17-positive ECs and progressors are not readily discernible using standard assays to measure immune responses.
Figures



Similar articles
-
Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication.J Virol. 2008 Feb;82(4):1723-38. doi: 10.1128/JVI.02084-07. Epub 2007 Dec 5. J Virol. 2008. PMID: 18057253 Free PMC article.
-
Vaccine-Induced Simian Immunodeficiency Virus-Specific CD8+ T-Cell Responses Focused on a Single Nef Epitope Select for Escape Variants Shortly after Infection.J Virol. 2015 Nov;89(21):10802-20. doi: 10.1128/JVI.01440-15. Epub 2015 Aug 19. J Virol. 2015. PMID: 26292326 Free PMC article.
-
Escape from CD8(+) T cell responses in Mamu-B*00801(+) macaques differentiates progressors from elite controllers.J Immunol. 2012 Apr 1;188(7):3364-70. doi: 10.4049/jimmunol.1102470. Epub 2012 Mar 2. J Immunol. 2012. PMID: 22387557 Free PMC article.
-
Mamu-B*17+ Rhesus Macaques Vaccinated with env, vif, and nef Manifest Early Control of SIVmac239 Replication.J Virol. 2018 Jul 31;92(16):e00690-18. doi: 10.1128/JVI.00690-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29875239 Free PMC article.
-
Infection with "escaped" virus variants impairs control of simian immunodeficiency virus SIVmac239 replication in Mamu-B*08-positive macaques.J Virol. 2009 Nov;83(22):11514-27. doi: 10.1128/JVI.01298-09. Epub 2009 Sep 2. J Virol. 2009. PMID: 19726517 Free PMC article.
Cited by
-
Ex vivo SIV-specific CD8 T cell responses in heterozygous animals are primarily directed against peptides presented by a single MHC haplotype.PLoS One. 2012;7(8):e43690. doi: 10.1371/journal.pone.0043690. Epub 2012 Aug 22. PLoS One. 2012. PMID: 22928016 Free PMC article.
-
HLA-B*14:02-Restricted Env-Specific CD8+ T-Cell Activity Has Highly Potent Antiviral Efficacy Associated with Immune Control of HIV Infection.J Virol. 2017 Oct 27;91(22):e00544-17. doi: 10.1128/JVI.00544-17. Print 2017 Nov 15. J Virol. 2017. PMID: 28878089 Free PMC article. Clinical Trial.
-
Is an HIV vaccine possible?Braz J Infect Dis. 2009 Aug;13(4):304-10. doi: 10.1590/s1413-86702009000400013. Braz J Infect Dis. 2009. PMID: 20231996 Free PMC article. Review.
-
Influence of the MHC genotype on the progression of experimental SIV infection in the Mauritian cynomolgus macaque.Immunogenetics. 2011 May;63(5):267-74. doi: 10.1007/s00251-010-0504-6. Epub 2011 Jan 14. Immunogenetics. 2011. PMID: 21234560
-
Immunopathogenesis of AIDS.Curr Infect Dis Rep. 2009 May;11(3):239-45. doi: 10.1007/s11908-009-0035-1. Curr Infect Dis Rep. 2009. PMID: 19366567
References
-
- Ali, A., S. Pillai, H. Ng, R. Lubong, D. D. Richman, B. D. Jamieson, Y. Ding, M. J. McElrath, J. C. Guatelli, and O. O. Yang. 2003. Broadly increased sensitivity to cytotoxic T lymphocytes resulting from Nef epitope escape mutations. J. Immunol. 1713999-4005. - PubMed
-
- Allen, T. M., M. Altfeld, S. C. Geer, E. T. Kalife, C. Moore, K. M. O'Sullivan, I. DeSouza, M. E. Feeney, R. L. Eldridge, E. L. Maier, D. E. Kaufmann, M. P. Lahaie, L. Reyor, G. Tanzi, M. N. Johnston, C. Brander, R. Draenert, J. K. Rockstroh, H. Jessen, E. S. Rosenberg, S. A. Mallal, and B. D. Walker. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J. Virol. 7913239-13249. - PMC - PubMed
-
- Allen, T. M., X. G. Yu, E. T. Kalife, L. L. Reyor, M. Lichterfeld, M. John, M. Cheng, R. L. Allgaier, S. Mui, N. Frahm, G. Alter, N. V. Brown, M. N. Johnston, E. S. Rosenberg, S. A. Mallal, C. Brander, B. D. Walker, and M. Altfeld. 2005. De novo generation of escape variant-specific CD8+ T-cell responses following cytotoxic T-lymphocyte escape in chronic human immunodeficiency virus type 1 infection. J. Virol. 7912952-12960. - PMC - PubMed
-
- Alter, G., M. P. Martin, N. Teigen, W. H. Carr, T. J. Suscovich, A. Schneidewind, H. Streeck, M. Waring, A. Meier, C. Brander, J. D. Lifson, T. M. Allen, M. Carrington, and M. Altfeld. 2007. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J. Exp. Med. 2043027-3036. - PMC - PubMed
-
- Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLoS Med. 3e403. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

