Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair
- PMID: 18385275
- PMCID: PMC2413152
- DOI: 10.1101/gr.075903.107
Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair
Abstract
Recurrent submicroscopic genomic copy number changes are the result of nonallelic homologous recombination (NAHR). Nonrecurrent aberrations, however, can result from different nonexclusive recombination-repair mechanisms. We previously described small microduplications at Xq28 containing MECP2 in four male patients with a severe neurological phenotype. Here, we report on the fine-mapping and breakpoint analysis of 16 unique microduplications. The size of the overlapping copy number changes varies between 0.3 and 2.3 Mb, and FISH analysis on three patients demonstrated a tandem orientation. Although eight of the 32 breakpoint regions coincide with low-copy repeats, none of the duplications are the result of NAHR. Bioinformatics analysis of the breakpoint regions demonstrated a 2.5-fold higher frequency of Alu interspersed repeats as compared with control regions, as well as a very high GC content (53%). Unexpectedly, we obtained the junction in only one patient by long-range PCR, which revealed nonhomologous end joining as the mechanism. Breakpoint analysis in two other patients by inverse PCR and subsequent array comparative genomic hybridization analysis demonstrated the presence of a second duplicated region more telomeric at Xq28, of which one copy was inserted in between the duplicated MECP2 regions. These data suggest a two-step mechanism in which part of Xq28 is first inserted near the MECP2 locus, followed by breakage-induced replication with strand invasion of the normal sister chromatid. Our results indicate that the mechanism by which copy number changes occur in regions with a complex genomic architecture can yield complex rearrangements.
Figures

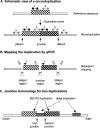


References
-
- Aradhya S., Woffendin H., Bonnen P., Heiss N.S., Yamagata T., Esposito T., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Woffendin H., Bonnen P., Heiss N.S., Yamagata T., Esposito T., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Bonnen P., Heiss N.S., Yamagata T., Esposito T., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Heiss N.S., Yamagata T., Esposito T., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Yamagata T., Esposito T., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Esposito T., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Bardaro T., Poustka A., D'Urso M., Kenwrick S., Poustka A., D'Urso M., Kenwrick S., D'Urso M., Kenwrick S., Kenwrick S., et al. Physical and genetic characterization reveals a pseudogene, an evolutionary junction, and unstable loci in distal Xq28. Genomics. 2002;79:31–40. - PubMed
-
- Balciuniene J., Feng N., Iyadurai K., Hirsch B., Charnas L., Bill B.R., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Feng N., Iyadurai K., Hirsch B., Charnas L., Bill B.R., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Iyadurai K., Hirsch B., Charnas L., Bill B.R., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Hirsch B., Charnas L., Bill B.R., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Charnas L., Bill B.R., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Bill B.R., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Easterday M.C., Staaf J., Oseth L., Czapansky-Beilman D., Staaf J., Oseth L., Czapansky-Beilman D., Oseth L., Czapansky-Beilman D., Czapansky-Beilman D., et al. Recurrent 10q22-q23 deletions: A genomic disorder on 10q associated with cognitive and behavioral abnormalities. Am. J. Hum. Genet. 2007;80:938–947. - PMC - PubMed
-
- Bauters M., Van Esch H., Marynen P., Froyen G., Van Esch H., Marynen P., Froyen G., Marynen P., Froyen G., Froyen G. X chromosome array-CGH for the identification of novel X-linked mental retardation genes. Eur. J. Med. Genet. 2005;48:263–275. - PubMed
-
- Deeb S.S., Kohl S., Kohl S. Genetics of color vision deficiencies. Dev. Ophthalmol. 2003;37:170–187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
