RAB-11 permissively regulates spindle alignment by modulating metaphase microtubule dynamics in Caenorhabditis elegans early embryos
- PMID: 18385514
- PMCID: PMC2397304
- DOI: 10.1091/mbc.e07-09-0862
RAB-11 permissively regulates spindle alignment by modulating metaphase microtubule dynamics in Caenorhabditis elegans early embryos
Abstract
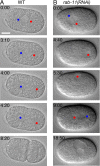
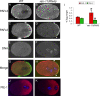
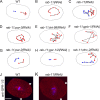
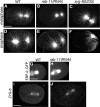
Alignment of the mitotic spindle along a preformed axis of polarity is crucial for generating cell diversity in many organisms, yet little is known about the role of the endomembrane system in this process. RAB-11 is a small GTPase enriched in recycling endosomes. When we depleted RAB-11 by RNAi in Caenorhabditis elegans, the spindle of the one-cell embryo failed to align along the axis of polarity in metaphase and underwent violent movements in anaphase. The distance between astral microtubules ends and the anterior cortex was significantly increased in rab-11(RNAi) embryos specifically during metaphase, possibly accounting for the observed spindle alignment defects. Additionally, we found that normal ER morphology requires functional RAB-11, particularly during metaphase. We hypothesize that RAB-11, in conjunction with the ER, acts to regulate cell cycle-specific changes in astral microtubule length to ensure proper spindle alignment in Caenorhabditis elegans early embryos.
Figures


Similar articles
-
RACK-1 directs dynactin-dependent RAB-11 endosomal recycling during mitosis in Caenorhabditis elegans.Mol Biol Cell. 2009 Mar;20(6):1629-38. doi: 10.1091/mbc.e08-09-0917. Epub 2009 Jan 21. Mol Biol Cell. 2009. PMID: 19158384 Free PMC article.
-
CLASPs function redundantly to regulate astral microtubules in the C. elegans embryo.Dev Biol. 2012 Aug 15;368(2):242-54. doi: 10.1016/j.ydbio.2012.05.016. Epub 2012 May 19. Dev Biol. 2012. PMID: 22613359 Free PMC article.
-
zyg-8, a gene required for spindle positioning in C. elegans, encodes a doublecortin-related kinase that promotes microtubule assembly.Dev Cell. 2001 Sep;1(3):363-75. doi: 10.1016/s1534-5807(01)00046-6. Dev Cell. 2001. PMID: 11702948
-
Spindle positioning during the asymmetric first cell division of Caenorhabditis elegans embryos.Novartis Found Symp. 2001;237:164-75; discussion 176-81. doi: 10.1002/0470846666.ch13. Novartis Found Symp. 2001. PMID: 11444042 Review.
-
Physical Limits on the Precision of Mitotic Spindle Positioning by Microtubule Pushing forces: Mechanics of mitotic spindle positioning.Bioessays. 2017 Nov;39(11):10.1002/bies.201700122. doi: 10.1002/bies.201700122. Epub 2017 Sep 28. Bioessays. 2017. PMID: 28960439 Free PMC article. Review.
Cited by
-
A new role for Rab GTPases during early mitotic stages.Small GTPases. 2014;5:e29565. doi: 10.4161/sgtp.29565. Epub 2014 Jun 12. Small GTPases. 2014. PMID: 24921241 Free PMC article.
-
RAB-5 controls the cortical organization and dynamics of PAR proteins to maintain C. elegans early embryonic polarity.PLoS One. 2012;7(4):e35286. doi: 10.1371/journal.pone.0035286. Epub 2012 Apr 24. PLoS One. 2012. PMID: 22545101 Free PMC article.
-
Loss of MYO5B expression deregulates late endosome size which hinders mitotic spindle orientation.PLoS Biol. 2019 Nov 4;17(11):e3000531. doi: 10.1371/journal.pbio.3000531. eCollection 2019 Nov. PLoS Biol. 2019. PMID: 31682603 Free PMC article.
-
From dusk till dawn: cell cycle progression in the red seaweed Gracilariopsis chorda (Rhodophyta).iScience. 2024 Jun 6;27(7):110190. doi: 10.1016/j.isci.2024.110190. eCollection 2024 Jul 19. iScience. 2024. PMID: 38984202 Free PMC article.
-
Profiling of the mammalian mitotic spindle proteome reveals an ER protein, OSTD-1, as being necessary for cell division and ER morphology.PLoS One. 2013 Oct 10;8(10):e77051. doi: 10.1371/journal.pone.0077051. eCollection 2013. PLoS One. 2013. PMID: 24130834 Free PMC article.
References
-
- Basham S. E., Rose L. S. Mutations in ooc-5 and ooc-3 disrupt oocyte formation and the reestablishment of asymmetric PAR protein localization in two-cell Caenorhabditis elegans embryos. Dev. Biol. 1999;215:253–263. - PubMed
-
- Basham S. E., Rose L. S. The Caenorhabditis elegans polarity gene ooc-5 encodes a Torsin-related protein of the AAA ATPase superfamily. Development. 2001;128:4645–4656. - PubMed
-
- Boyd L., Guo S., Levitan D., Stinchcomb D. T., Kemphues K. J. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development. 1996;122:3075–3084. - PubMed
-
- Capes-Davis A., Tolhurst O., Dunn J. M., Jeffrey P. L. Expression of doublecortin (DCX) and doublecortin-like kinase (DCLK) within the developing chick brain. Dev. Dyn. 2005;232:457–467. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

