Translating insights from the cancer genome into clinical practice
- PMID: 18385729
- PMCID: PMC2730524
- DOI: 10.1038/nature06914
Translating insights from the cancer genome into clinical practice
Abstract
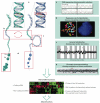
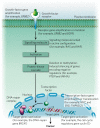
Cancer cells have diverse biological capabilities that are conferred by numerous genetic aberrations and epigenetic modifications. Today's powerful technologies are enabling these changes to the genome to be catalogued in detail. Tomorrow is likely to bring a complete atlas of the reversible and irreversible alterations that occur in individual cancers. The challenge now is to work out which molecular abnormalities contribute to cancer and which are simply 'noise' at the genomic and epigenomic levels. Distinguishing between these will aid in understanding how the aberrations in a cancer cell collaborate to drive pathophysiology. Past successes in converting information from genomic discoveries into clinical tools provide valuable lessons to guide the translation of emerging insights from the genome into clinical end points that can affect the practice of cancer medicine.
Figures

References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci. Am. 2007;296:50–57. - PubMed
-
- Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. - PubMed
-
- Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. - PubMed
-
-
Pao W, et al. EGF receptor gene mutations are common in lung cancers from ‘never smokers’ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl Acad. Sci. USA. 2004;101:13306–13311.References -5 show that a subset of patients with lung cancer have EGFR mutations and are responsive to an EGFR-specific tyrosine kinase inhibitor, a finding based on prospective analyses of retrospective data.
-
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

