NAD(P)H quinone oxidoreductase 1 inhibits the proteasomal degradation of the tumour suppressor p33(ING1b)
- PMID: 18388957
- PMCID: PMC2427386
- DOI: 10.1038/embor.2008.48
NAD(P)H quinone oxidoreductase 1 inhibits the proteasomal degradation of the tumour suppressor p33(ING1b)
Abstract
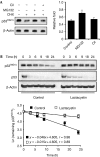
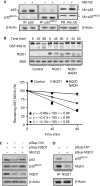
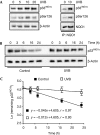
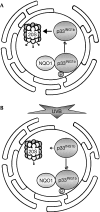
The tumour suppressor p33(ING1b) ((ING1b) for inhibitor of growth family, member 1b) is important in cellular stress responses, including cell-cycle arrest, apoptosis, chromatin remodelling and DNA repair; however, its degradation pathway is still unknown. Recently, we showed that genotoxic stress induces p33(ING1b) phosphorylation at Ser 126, and abolishment of Ser 126 phosphorylation markedly shortened its half-life. Therefore, we suggest that Ser 126 phosphorylation modulates the interaction of p33(ING1b) with its degradation machinery, stabilizing this protein. Combining the use of inhibitors of the main degradation pathways in the nucleus (proteasome and calpains), partial isolation of the proteasome complex, and in vitro interaction and degradation assays, we set out to determine the degradation mechanism of p33(ING1b). We found that p33(ING1b) is degraded in the 20S proteasome and that NAD(P)H quinone oxidoreductase 1 (NQO1), an oxidoreductase previously shown to modulate the degradation of p53 in the 20S proteasome, inhibits the degradation of p33(ING1b). Furthermore, ultraviolet irradiation induces p33(ING1b) phosphorylation at Ser 126, which, in turn, facilitates its interaction with NQO1.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Phosphorylation of the tumor suppressor p33(ING1b) at Ser-126 influences its protein stability and proliferation of melanoma cells.FASEB J. 2007 Nov;21(13):3705-16. doi: 10.1096/fj.07-8069com. Epub 2007 Jun 21. FASEB J. 2007. PMID: 17585055
-
Adenovirus-mediated expression of p33(ING1b) induces apoptosis and inhibits proliferation in gastric adenocarcinoma cells in vitro.Gastric Cancer. 2012 Oct;15(4):355-62. doi: 10.1007/s10120-011-0123-4. Epub 2012 Jan 12. Gastric Cancer. 2012. PMID: 22237655
-
Nuclear to cytoplasmic shift of p33(ING1b) protein from normal oral mucosa to oral squamous cell carcinoma in relation to clinicopathological variables.J Cancer Res Clin Oncol. 2008 Mar;134(3):421-6. doi: 10.1007/s00432-007-0305-y. Epub 2007 Sep 6. J Cancer Res Clin Oncol. 2008. PMID: 17805569 Free PMC article.
-
NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector.Arch Biochem Biophys. 2010 Sep 1;501(1):116-23. doi: 10.1016/j.abb.2010.03.019. Epub 2010 Mar 31. Arch Biochem Biophys. 2010. PMID: 20361926 Free PMC article. Review.
-
The role of the tumour suppressor p33 ING1b in human neoplasia.J Clin Pathol. 2003 Jul;56(7):491-6. doi: 10.1136/jcp.56.7.491. J Clin Pathol. 2003. PMID: 12835293 Free PMC article. Review.
Cited by
-
The diverse functionality of NQO1 and its roles in redox control.Redox Biol. 2021 May;41:101950. doi: 10.1016/j.redox.2021.101950. Epub 2021 Mar 20. Redox Biol. 2021. PMID: 33774477 Free PMC article. Review.
-
NQO1 potentiates apoptosis evasion and upregulates XIAP via inhibiting proteasome-mediated degradation SIRT6 in hepatocellular carcinoma.Cell Commun Signal. 2019 Dec 16;17(1):168. doi: 10.1186/s12964-019-0491-7. Cell Commun Signal. 2019. PMID: 31842909 Free PMC article.
-
Roles of NAD(P)H:quinone Oxidoreductase 1 in Diverse Diseases.Life (Basel). 2021 Nov 26;11(12):1301. doi: 10.3390/life11121301. Life (Basel). 2021. PMID: 34947831 Free PMC article. Review.
-
The NQO1*2/*2 polymorphism is associated with poor overall survival in patients following resection of stages II and IIIa non-small cell lung cancer.Oncol Rep. 2011 Jun;25(6):1765-72. doi: 10.3892/or.2011.1249. Epub 2011 Apr 6. Oncol Rep. 2011. PMID: 21479364 Free PMC article.
-
The protein level of PGC-1α, a key metabolic regulator, is controlled by NADH-NQO1.Mol Cell Biol. 2013 Jul;33(13):2603-13. doi: 10.1128/MCB.01672-12. Epub 2013 May 6. Mol Cell Biol. 2013. PMID: 23648480 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

