Evolution of a TRIM5-CypA splice isoform in old world monkeys
- PMID: 18389077
- PMCID: PMC2279257
- DOI: 10.1371/journal.ppat.1000003
Evolution of a TRIM5-CypA splice isoform in old world monkeys
Abstract
The TRIM family proteins share a conserved arrangement of three adjacent domains, an N-terminal RING domain, followed by one or two B-boxes and a coiled-coil, which constitutes the tripartite-motif for which the family is named. However, the C-termini of TRIM proteins vary, and include at least nine evolutionarily distinct, unrelated protein domains. Antiviral restriction factor TRIM5alpha has a C-terminal B30.2/SPRY domain, which is the major determinant of viral target specificity. Here, we describe the evolution of a cyclophilin-A encoding exon downstream of the TRIM5 locus of Asian macaques. Alternative splicing gives rise to chimeric transcripts encoding the TRIM motif fused to a C-terminal CypA domain (TRIM5-CypA). We detected TRIM5-CypA chimeric transcripts in primary lymphocytes from two macaque species. These were derived in part from a CypA pseudogene in the TRIM5 locus, which is distinct from the previously described CypA insertion in TRIM5 of owl monkeys. The CypA insertion is linked to a mutation in the 3' splice site upstream of exon 7, which may prevent or reduce expression of the alpha-isoform. All pig-tailed macaques (M. nemestrina) screened were homozygous for the CypA insertion. In contrast, the CypA-containing allele was present in 17% (17/101) of rhesus macaques (M. mulatta). The block to HIV-1 infection in lymphocytes from animals bearing the TRIM5-CypA allele was weaker than that in cells from wild type animals. HIV-1 infectivity remained significantly lower than SIV infectivity, but was not rescued by treatment with cyclosporine A. Thus, unlike owl monkey TRIMCyp, expression of the macaque TRIM5-CypA isoform does not result in increased restriction of HIV-1. Despite its distinct evolutionary origin, Macaca TRIM5-CypA has a similar domain arrangement and shares approximately 80% amino-acid identity with the TRIMCyp protein of owl monkeys. The independent appearance of TRIM5-CypA chimeras in two primate lineages constitutes a remarkable example of convergent evolution. Based on the presence of the CypA insertion in separate macaque lineages, and its absence from sooty mangabeys, we estimate that the Macaca TRIM5-CypA variant appeared 5-10 million years ago in a common ancestor of the Asian macaques. Whether the formation of novel genes through alternative splicing has played a wider role in the evolution of the TRIM family remains to be investigated.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
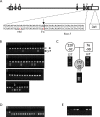
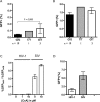


References
-
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

