Targeted disruption of the basic Krüppel-like factor gene (Klf3) reveals a role in adipogenesis
- PMID: 18391014
- PMCID: PMC2423134
- DOI: 10.1128/MCB.01942-07
Targeted disruption of the basic Krüppel-like factor gene (Klf3) reveals a role in adipogenesis
Abstract
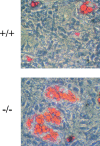
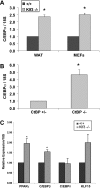
Krüppel-like factors (KLFs) recognize CACCC and GC-rich sequences in gene regulatory elements. Here, we describe the disruption of the murine basic Krüppel-like factor gene (Bklf or Klf3). Klf3 knockout mice have less white adipose tissue, and their fat pads contain smaller and fewer cells. Adipocyte differentiation is altered in murine embryonic fibroblasts from Klf3 knockouts. Klf3 expression was studied in the 3T3-L1 cellular system. Adipocyte differentiation is accompanied by a decline in Klf3 expression, and forced overexpression of Klf3 blocks 3T3-L1 differentiation. Klf3 represses transcription by recruiting C-terminal binding protein (CtBP) corepressors. CtBPs bind NADH and may function as metabolic sensors. A Klf3 mutant that does not bind CtBP cannot block adipogenesis. Other KLFs, Klf2, Klf5, and Klf15, also regulate adipogenesis, and functional CACCC elements occur in key adipogenic genes, including in the C/ebpalpha promoter. We find that C/ebpalpha is derepressed in Klf3 and Ctbp knockout fibroblasts and adipocytes from Klf3 knockout mice. Chromatin immunoprecipitations confirm that Klf3 binds the C/ebpalpha promoter in vivo. These results implicate Klf3 and CtBP in controlling adipogenesis.
Figures





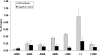
References
-
- Balasubramanian, P., L.-J. Zhao, and G. Chinnadurai. 2003. Nicotinamide adenine dinucleotide stimulates oligomerization, interaction with adenovirus E1A and an intrinsic dehydrogenase activity of CtBP. FEBS Lett. 537157-160. - PubMed
-
- Banerjee, S. S., M. W. Feinberg, M. Watanabe, S. Gray, R. L. Haspel, D. J. Denkinger, R. Kawahara, H. Hauner, and M. K. Jain. 2003. The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J. Biol. Chem. 2782581-2584. - PubMed
-
- Bieker, J. J. 1996. Isolation, genomic structure, and expression of human erythroid Kruppel-like factor (EKLF). DNA Cell Biol. 15347-352. - PubMed
-
- Bieker, J. J. 2001. Kruppel-like factors: three fingers in many pies. J. Biol. Chem. 27634355-34358. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
