Specific effects of KChIP3/calsenilin/DREAM, but not KChIPs 1, 2 and 4, on calcium signalling and regulated secretion in PC12 cells
- PMID: 18393943
- PMCID: PMC2474559
- DOI: 10.1042/BJ20080441
Specific effects of KChIP3/calsenilin/DREAM, but not KChIPs 1, 2 and 4, on calcium signalling and regulated secretion in PC12 cells
Abstract
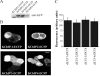
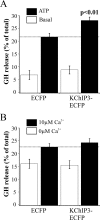
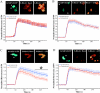
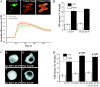
The KChIPs (K+ channel-interacting proteins) are members of the NCS (neuronal calcium sensor) protein family of Ca2+-binding proteins. It is unclear to what extent the KChIPs have distinct functions although they all interact with Kv4 K+ channels. KChIP3 has also been shown to repress transcription of specific genes via binding to DRE (downstream regulatory element) motifs and all KChIPs may share this function. In the present study, we have compared the function of isoforms of the four KChIPs. KChIPs 1-4 were found to stimulate the traffic of Kv4.2 channels to the plasma membrane. KChIP3 expression in PC12 cells resulted in an increase in exocytosis evoked by activation of purinergic receptors. In contrast, KChIPs 1, 2 and 4, although expressed to the same extent, had no effect on secretion. In addition, KChIP3 but not KChIPs 1, 2 and 4 modified the ATP-induced Ca2+ signal resulting in a delay in recovery after the peak Ca2+ elevation and also specifically resulted in down-regulation of the Na+/Ca2+ exchanger NCX3, which could explain the effects on the Ca2+ signal and secretion. Regulation of NCX3 by KChIP3 has been shown to occur via its DREAM (DRE antagonist modulator) function [Gomez-Villafuertes, Torres, Barrio, Savignac, Gabellini, Rizzato, Pintado, Gutierrez-Adan, Mellstrom, Carafoli and Naranjo (2005) J. Neurosci. 25, 10822-10830] suggesting that this activity might depend on the cellular context of expression of the various KChIPs. These results reveal a new role for KChIP3 in the regulation of Ca2+-regulated secretion and also suggest that the functions of each of the KChIPs may be more specialized than previously appreciated.
Figures


References
-
- Palczewski K., Polans A., Baehr W., Ames J. B. Ca2+-binding proteins in the retina: structure, function and the etiology of human visual diseases. BioEssays. 2000;22:337–350. - PubMed
-
- Pruunsild P., Timmusk T. Structure, alternative splicing, and expression of the human and mouse KCNIP gene family. Genomics. 2005;86:581–593. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

